Inferring the Chemotactic Strategy of P. putida and E. coli Using Modified Kramers-Moyal Coefficients
- PMID: 28114420
- PMCID: PMC5293273
- DOI: 10.1371/journal.pcbi.1005329
Inferring the Chemotactic Strategy of P. putida and E. coli Using Modified Kramers-Moyal Coefficients
Abstract
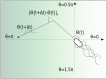
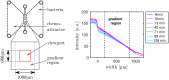
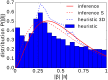
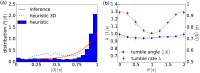
Many bacteria perform a run-and-tumble random walk to explore their surrounding and to perform chemotaxis. In this article we present a novel method to infer the relevant parameters of bacterial motion from experimental trajectories including the tumbling events. We introduce a stochastic model for the orientation angle, where a shot-noise process initiates tumbles, and analytically calculate conditional moments, reminiscent of Kramers-Moyal coefficients. Matching them with the moments calculated from experimental trajectories of the bacteria E. coli and Pseudomonas putida, we are able to infer their respective tumble rates, the rotational diffusion constants, and the distributions of tumble angles in good agreement with results from conventional tumble recognizers. We also define a novel tumble recognizer, which explicitly quantifies the error in recognizing tumbles. In the presence of a chemical gradient we condition the moments on the bacterial direction of motion and thereby explore the chemotaxis strategy. For both bacteria we recover and quantify the classical chemotactic strategy, where the tumble rate is smallest along the chemical gradient. In addition, for E. coli we detect some cells, which bias their mean tumble angle towards smaller values. Our findings are supported by a scaling analysis of appropriate ratios of conditional moments, which are directly calculated from experimental data.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

