A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents
- PMID: 28114422
- PMCID: PMC5256994
- DOI: 10.1371/journal.pone.0170628
A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents
Abstract
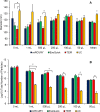
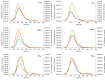
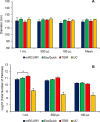
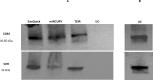
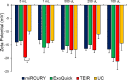
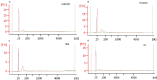
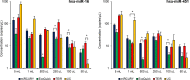
Exosomes play a role in cell-to-cell signaling and serve as possible biomarkers. Isolating exosomes with reliable quality and substantial concentration is a major challenge. Our purpose is to compare the exosomes extracted by three different exosome isolation kits (miRCURY, ExoQuick, and Invitrogen Total Exosome Isolation Reagent) and differential ultracentrifugation (UC) using six different volumes of a non-cancerous human serum (5 ml, 1 ml, 500 μl, 250 μl, 100 μl, and 50 μl) and three different volumes (1 ml, 500 μl and 100 μl) of six individual commercial serum samples collected from human donors. The smaller starting volumes (100 μl and 50 μl) are used to mimic conditions of limited availability of heterogeneous biological samples. The isolated exosomes were characterized based upon size, quantity, zeta potential, CD63 and CD9 protein expression, and exosomal RNA (exRNA) quality and quantity using several complementary methods: nanoparticle tracking analysis (NTA) with ZetaView, western blot, transmission electron microscopy (TEM), the Agilent Bioanalyzer system, and droplet digital PCR (ddPCR). Our NTA results showed that all isolation techniques produced exosomes within the expected size range (40-150 nm). The three kits, though, produced a significantly higher yield (80-300 fold) of exosomes as compared to UC for all serum volumes, except 5 mL. We also found that exosomes isolated by the different techniques and serum volumes had similar zeta potentials to previous studies. Western blot analysis and TEM immunogold labelling confirmed the expression of two common exosomal protein markers, CD63 and CD9, in samples isolated by all techniques. All exosome isolations yielded high quality exRNA, containing mostly small RNA with a peak between 25 and 200 nucleotides in size. ddPCR results indicated that exosomes isolated from similar serum volumes but different isolation techniques rendered similar concentrations of two selected exRNA: hsa-miR-16 and hsa-miR-451. In summary, the three commercial exosome isolation kits are viable alternatives to UC, even when limited amounts of biological samples are available.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, et al. Exosomes with immune modulatory features are present in human breast milk. Journal of immunology. 2007;179(3):1969–78. - PubMed
-
- Alegre E, Sanmamed MF, Rodriguez C, Carranza O, Martin-Algarra S, Gonzalez A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Archives of pathology & laboratory medicine. 2014;138(6):828–32. - PubMed
-
- Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. British journal of cancer. 2009;100(10):1603–7. PubMed Central PMCID: PMC2696767. 10.1038/sj.bjc.6605058 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

