Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing
- PMID: 28115465
- PMCID: PMC5287113
- DOI: 10.1101/gad.286963.116
Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing
Abstract
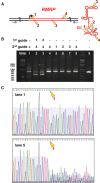
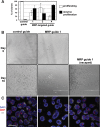
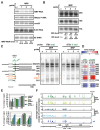
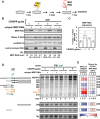
MRP RNA is an abundant, essential noncoding RNA whose functions have been proposed in yeast but are incompletely understood in humans. Mutations in the genomic locus for MRP RNA cause pleiotropic human diseases, including cartilage hair hypoplasia (CHH). Here we applied CRISPR-Cas9 genome editing to disrupt the endogenous human MRP RNA locus, thereby attaining what has eluded RNAi and RNase H experiments: elimination of MRP RNA in the majority of cells. The resulting accumulation of ribosomal RNA (rRNA) precursor-analyzed by RNA fluorescent in situ hybridization (FISH), Northern blots, and RNA sequencing-implicates MRP RNA in pre-rRNA processing. Amelioration of pre-rRNA imbalance is achieved through rescue of MRP RNA levels by ectopic expression. Furthermore, affinity-purified MRP ribonucleoprotein (RNP) from HeLa cells cleaves the human pre-rRNA in vitro at at least one site used in cells, while RNP isolated from cells with CRISPR-edited MRP loci loses this activity, and ectopic MRP RNA expression restores cleavage activity. Thus, a role for RNase MRP in human pre-rRNA processing is established. As demonstrated here, targeted CRISPR disruption is a valuable tool for functional studies of essential noncoding RNAs that are resistant to RNAi and RNase H-based degradation.
Keywords: CRISPR–Cas9; RMRP; RNase MRP; RNase P; preribosomal RNA processing.
© 2017 Goldfarb and Cech; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Altman S, Smith JD. 1971. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol 233: 35–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
