A semisynthetic organism engineered for the stable expansion of the genetic alphabet
- PMID: 28115716
- PMCID: PMC5307467
- DOI: 10.1073/pnas.1616443114
A semisynthetic organism engineered for the stable expansion of the genetic alphabet
Abstract
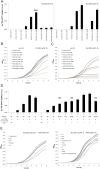
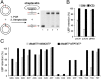
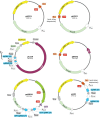
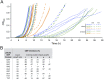
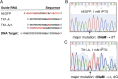
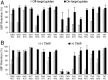
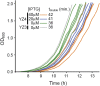
All natural organisms store genetic information in a four-letter, two-base-pair genetic alphabet. The expansion of the genetic alphabet with two synthetic unnatural nucleotides that selectively pair to form an unnatural base pair (UBP) would increase the information storage potential of DNA, and semisynthetic organisms (SSOs) that stably harbor this expanded alphabet would thereby have the potential to store and retrieve increased information. Toward this goal, we previously reported that Escherichia coli grown in the presence of the unnatural nucleoside triphosphates dNaMTP and d5SICSTP, and provided with the means to import them via expression of a plasmid-borne nucleoside triphosphate transporter, replicates DNA containing a single dNaM-d5SICS UBP. Although this represented an important proof-of-concept, the nascent SSO grew poorly and, more problematically, required growth under controlled conditions and even then was unable to indefinitely store the unnatural information, which is clearly a prerequisite for true semisynthetic life. Here, to fortify and vivify the nascent SSO, we engineered the transporter, used a more chemically optimized UBP, and harnessed the power of the bacterial immune response by using Cas9 to eliminate DNA that had lost the UBP. The optimized SSO grows robustly, constitutively imports the unnatural triphosphates, and is able to indefinitely retain multiple UBPs in virtually any sequence context. This SSO is thus a form of life that can stably store genetic information using a six-letter, three-base-pair alphabet.
Keywords: CRISPR; Cas9; DNA replication; nucleotide transporter; unnatural base pair.
Conflict of interest statement
Y.Z., B.M.L., and F.E.R. have filed a patent application based on the use of Cas9 for enforced retention of the UBP. Y.Z. and F.E.R. have filed a patent application for the truncated transporter. F.E.R. has a financial interest (shares) in Synthorx Inc., a company that has commercial interests in the UBP. The other authors declare no other competing financial interests.
Figures





Comment in
-
Reply to Hettinger: Hydrophobic unnatural base pairs and the expansion of the genetic alphabet.Proc Natl Acad Sci U S A. 2017 Aug 8;114(32):E6478-E6479. doi: 10.1073/pnas.1708259114. Epub 2017 Aug 2. Proc Natl Acad Sci U S A. 2017. PMID: 28768812 Free PMC article. No abstract available.
-
Helix instability and self-pairing prevent unnatural base pairs from expanding the genetic alphabet.Proc Natl Acad Sci U S A. 2017 Aug 8;114(32):E6476-E6477. doi: 10.1073/pnas.1703423114. Epub 2017 Aug 2. Proc Natl Acad Sci U S A. 2017. PMID: 28768813 Free PMC article. No abstract available.
References
-
- Kimoto M, Hirao I. 2014. Creation of unnatural base pair systems toward new DNA/RNA biotechnologies. Chemical Biology of Nucleic Acids: Fundamentals and Clinical Applications, eds Erdmann AV, Markiewicz TW, Barciszewski J (Springer, Berlin), pp 131–148.
-
- Leduc S. The Mechanisms of Life. Rebman; New York: 1911.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

