Sex-specific chromatin landscapes in an ultra-compact chordate genome
- PMID: 28115992
- PMCID: PMC5240408
- DOI: 10.1186/s13072-016-0110-4
Sex-specific chromatin landscapes in an ultra-compact chordate genome
Abstract
Background: In multicellular organisms, epigenome dynamics are associated with transitions in the cell cycle, development, germline specification, gametogenesis and inheritance. Evolutionarily, regulatory space has increased in complex metazoans to accommodate these functions. In tunicates, the sister lineage to vertebrates, we examine epigenome adaptations to strong secondary genome compaction, sex chromosome evolution and cell cycle modes.
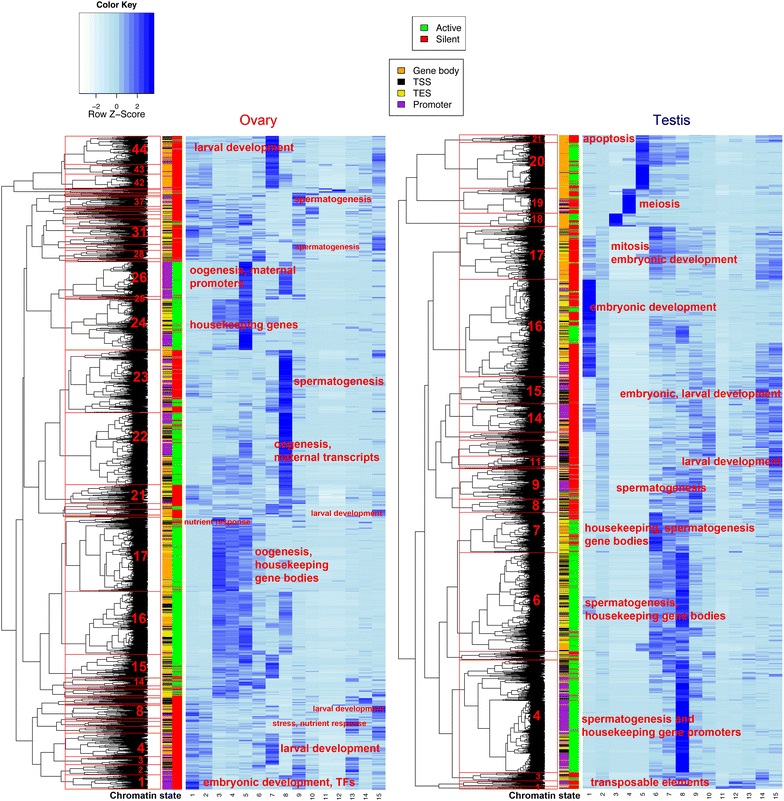
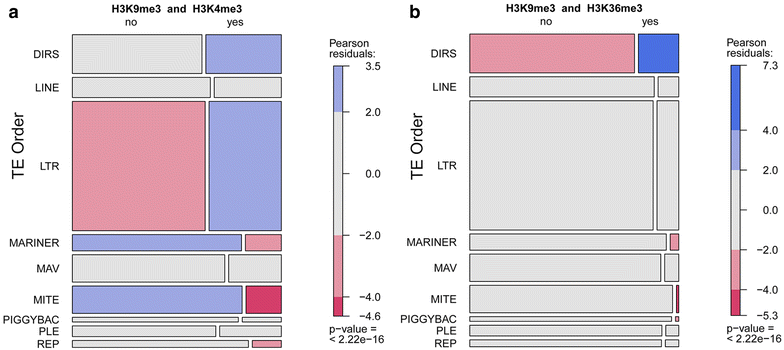
Results: Across the 70 MB Oikopleura dioica genome, we profiled 19 histone modifications, and RNA polymerase II, CTCF and p300 occupancies, to define chromatin states within two homogeneous tissues with distinct cell cycle modes: ovarian endocycling nurse nuclei and mitotically proliferating germ nuclei in testes. Nurse nuclei had active chromatin states similar to other metazoan epigenomes, with large domains of operon-associated transcription, a general lack of heterochromatin, and a possible role of Polycomb PRC2 in dosage compensation. Testis chromatin states reflected transcriptional activity linked to spermatogenesis and epigenetic marks that have been associated with establishment of transgenerational inheritance in other organisms. We also uncovered an unusual chromatin state specific to the Y-chromosome, which combined active and heterochromatic histone modifications on specific transposable elements classes, perhaps involved in regulating their activity.
Conclusions: Compacted regulatory space in this tunicate genome is accompanied by reduced heterochromatin and chromatin state domain widths. Enhancers, promoters and protein-coding genes have conserved epigenomic features, with adaptations to the organization of a proportion of genes in operon units. We further identified features specific to sex chromosomes, cell cycle modes, germline identity and dosage compensation, and unusual combinations of histone PTMs with opposing consensus functions.
Keywords: Dosage compensation; Endocycle; Enhancer; Heterochromatin; Histone; Polycomb; Spermatogenesis; Transposable elements.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

