Overexpression of PCNA Attenuates Oxidative Stress-Caused Delay of Gap-Filling during Repair of UV-Induced DNA Damage
- PMID: 28116145
- PMCID: PMC5237465
- DOI: 10.1155/2017/8154646
Overexpression of PCNA Attenuates Oxidative Stress-Caused Delay of Gap-Filling during Repair of UV-Induced DNA Damage
Abstract
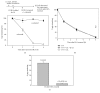
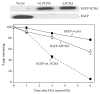
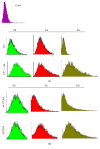
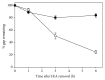
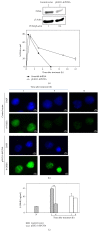
UVC irradiation-caused DNA lesions are repaired in mammalian cells solely by nucleotide excision repair (NER), which consists of sequential events including initial damage recognition, dual incision of damage site, gap-filling, and ligation. We have previously shown that gap-filling during the repair of UV-induced DNA lesions may be delayed by a subsequent treatment of oxidants or prooxidants such as hydrogen peroxide, flavonoids, and colcemid. We considered the delay as a result of competition for limiting protein/enzyme factor(s) during repair synthesis between NER and base excision repair (BER) induced by the oxidative chemicals. In this report, using colcemid as oxidative stress inducer, we showed that colcemid-caused delay of gap-filling during the repair of UV-induced DNA lesions was attenuated by overexpression of PCNA but not ligase-I. PCNA knockdown, as expected, delayed the gap-filling of NER but also impaired the repair of oxidative DNA damage. Fen-1 knockdown, however, did not affect the repair of oxidative DNA damage, suggesting repair of oxidative DNA damage is not of long patch BER. Furthermore, overexpression of XRCC1 delayed the gap-filling, and presumably increase of XRCC1 pulls PCNA away from gap-filling of NER for BER, consistent with our hypothesis that delay of gap-filling of NER attributes the competition between NER and BER.
Conflict of interest statement
The authors declare that there is no conflict of interests regarding the publication of this paper.
Figures



Similar articles
-
Delay of gap filling during nucleotide excision repair by base excision repair: the concept of competition exemplified by the effect of propolis.Toxicol Sci. 2011 Aug;122(2):339-48. doi: 10.1093/toxsci/kfr107. Epub 2011 May 10. Toxicol Sci. 2011. PMID: 21561884
-
Differential DNA damage response to UV and hydrogen peroxide depending of differentiation stage in a neuroblastoma model.Neurotoxicology. 2012 Oct;33(5):1086-95. doi: 10.1016/j.neuro.2012.05.017. Epub 2012 Jun 9. Neurotoxicology. 2012. PMID: 22687812
-
Resistance of human nucleotide excision repair synthesis in vitro to p21Cdn1.Oncogene. 1998 Dec 3;17(22):2827-38. doi: 10.1038/sj.onc.1202352. Oncogene. 1998. PMID: 9879989
-
Excision of Oxidatively Generated Guanine Lesions by Competitive DNA Repair Pathways.Int J Mol Sci. 2021 Mar 7;22(5):2698. doi: 10.3390/ijms22052698. Int J Mol Sci. 2021. PMID: 33800059 Free PMC article. Review.
-
Reprint of "Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair".DNA Repair (Amst). 2015 Dec;36:86-90. doi: 10.1016/j.dnarep.2015.11.002. Epub 2015 Nov 12. DNA Repair (Amst). 2015. PMID: 26596511 Free PMC article. Review.
Cited by
-
Programming of Cell Resistance to Genotoxic and Oxidative Stress.Biomedicines. 2018 Jan 2;6(1):5. doi: 10.3390/biomedicines6010005. Biomedicines. 2018. PMID: 29301323 Free PMC article. Review.
-
A FRET-Based Assay for the Identification of PCNA Inhibitors.Int J Mol Sci. 2023 Jul 24;24(14):11858. doi: 10.3390/ijms241411858. Int J Mol Sci. 2023. PMID: 37511614 Free PMC article.
-
Exploring the Antiproliferative and Modulatory Effects of 1-Methoxyisobrassinin on Ovarian Cancer Cells: Insights into Cell Cycle Regulation, Apoptosis, Autophagy, and Its Interactions with NAC.Molecules. 2024 Apr 13;29(8):1773. doi: 10.3390/molecules29081773. Molecules. 2024. PMID: 38675591 Free PMC article.
-
Within species expressed genetic variability and gene expression response to different temperatures in the rotifer Brachionus calyciflorus sensu stricto.PLoS One. 2019 Sep 30;14(9):e0223134. doi: 10.1371/journal.pone.0223134. eCollection 2019. PLoS One. 2019. PMID: 31568501 Free PMC article.
References
-
- Friedberg E. C. DNA Repair and Mutagenesis. 2nd. Washington, DC, USA: ASM Press; 2006.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

