Accelerating the search for the missing proteins in the human proteome
- PMID: 28117396
- PMCID: PMC5286205
- DOI: 10.1038/ncomms14271
Accelerating the search for the missing proteins in the human proteome
Abstract
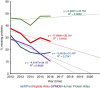
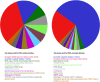
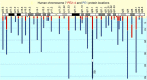
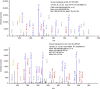
The Human Proteome Project (HPP) aims to discover high-stringency data for all proteins encoded by the human genome. Currently, ∼18% of the proteins in the human proteome (the missing proteins) do not have high-stringency evidence (for example, mass spectrometry) confirming their existence, while much additional information is available about many of these missing proteins. Here, we present MissingProteinPedia as a community resource to accelerate the discovery and understanding of these missing proteins.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Paik Y. K. et al.. The Chromosome-centric Human Proteome Project for cataloging proteins encoded in the genome. Nat. Biotechnol. 30, 221–223 (2012) Aims to define full set of human proteins encoded by ∼20,300 genes, chromosome-by-chromosome including tissue localization, isoforms and PTMs using MS and Abs. First coined term ‘missing proteins'. - PubMed
-
- Paik Y. K. et al.. Standard guidelines for the Chromosome-centric Human Proteome Project. J. Proteome Res. 11, 2005–2013 (2012). - PubMed
-
- Omenn G. S. et al.. Metrics for the Human Proteome Project 2016: progress on identifying and characterizing the human proteome, including post-translational modifications. J. Proteome Res. 15, 3951–3960 (2016) Update on HPP annual communal data re-analyses that adopted higher stringency MS metrics for protein evidence (PE1 = two unitypic peptides > 9 residues). HPP (neXtProt version 2016-02) has 16,518 PE1 proteins, with 2,949 PE2-4 missing proteins and 485 reclassified by higher stringency HPP Guidelines v2.0 to reduce false positives. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

