GSNFS: Gene subnetwork biomarker identification of lung cancer expression data
- PMID: 28117655
- PMCID: PMC5260788
- DOI: 10.1186/s12920-016-0231-4
GSNFS: Gene subnetwork biomarker identification of lung cancer expression data
Abstract
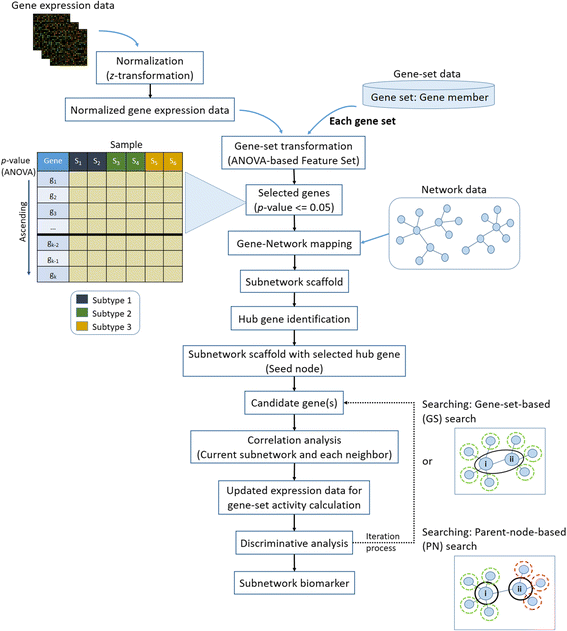
Background: Gene expression has been used to identify disease gene biomarkers, but there are ongoing challenges. Single gene or gene-set biomarkers are inadequate to provide sufficient understanding of complex disease mechanisms and the relationship among those genes. Network-based methods have thus been considered for inferring the interaction within a group of genes to further study the disease mechanism. Recently, the Gene-Network-based Feature Set (GNFS), which is capable of handling case-control and multiclass expression for gene biomarker identification, has been proposed, partly taking into account of network topology. However, its performance relies on a greedy search for building subnetworks and thus requires further improvement. In this work, we establish a new approach named Gene Sub-Network-based Feature Selection (GSNFS) by implementing the GNFS framework with two proposed searching and scoring algorithms, namely gene-set-based (GS) search and parent-node-based (PN) search, to identify subnetworks. An additional dataset is used to validate the results.
Methods: The two proposed searching algorithms of the GSNFS method for subnetwork expansion are concerned with the degree of connectivity and the scoring scheme for building subnetworks and their topology. For each iteration of expansion, the neighbour genes of a current subnetwork, whose expression data improved the overall subnetwork score, is recruited. While the GS search calculated the subnetwork score using an activity score of a current subnetwork and the gene expression values of its neighbours, the PN search uses the expression value of the corresponding parent of each neighbour gene. Four lung cancer expression datasets were used for subnetwork identification. In addition, using pathway data and protein-protein interaction as network data in order to consider the interaction among significant genes were discussed. Classification was performed to compare the performance of the identified gene subnetworks with three subnetwork identification algorithms.
Results: The two searching algorithms resulted in better classification and gene/gene-set agreement compared to the original greedy search of the GNFS method. The identified lung cancer subnetwork using the proposed searching algorithm resulted in an improvement of the cross-dataset validation and an increase in the consistency of findings between two independent datasets. The homogeneity measurement of the datasets was conducted to assess dataset compatibility in cross-dataset validation. The lung cancer dataset with higher homogeneity showed a better result when using the GS search while the dataset with low homogeneity showed a better result when using the PN search. The 10-fold cross-dataset validation on the independent lung cancer datasets showed higher classification performance of the proposed algorithms when compared with the greedy search in the original GNFS method.
Conclusions: The proposed searching algorithms provide a higher number of genes in the subnetwork expansion step than the greedy algorithm. As a result, the performance of the subnetworks identified from the GSNFS method was improved in terms of classification performance and gene/gene-set level agreement depending on the homogeneity of the datasets used in the analysis. Some common genes obtained from the four datasets using different searching algorithms are genes known to play a role in lung cancer. The improvement of classification performance and the gene/gene-set level agreement, and the biological relevance indicated the effectiveness of the GSNFS method for gene subnetwork identification using expression data.
Figures
References
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

