Enhanced Nonenzymatic RNA Copying with 2-Aminoimidazole Activated Nucleotides
- PMID: 28117989
- PMCID: PMC6326525
- DOI: 10.1021/jacs.6b13148
Enhanced Nonenzymatic RNA Copying with 2-Aminoimidazole Activated Nucleotides
Abstract
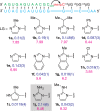
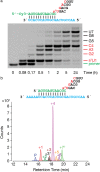
Achieving efficient nonenzymatic replication of RNA is an important step toward the synthesis of self-replicating protocells that may mimic early forms of life. Despite recent progress, the nonenzymatic copying of templates containing mixed sequences remains slow and inefficient. Here we demonstrate that activating nucleotides with 2-aminoimidazole results in superior reaction kinetics and improved yields of primer extension reaction products. This new leaving group significantly accelerates monomer addition as well as trimer-assisted RNA primer extension, allowing efficient copying of a variety of short RNA templates with mixed sequences.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Inoue T.; Orgel L. E. J. Am. Chem. Soc. 1981, 103, 7666–7667. 10.1021/ja00415a051. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

