Genome-Wide Association between Transcription Factor Expression and Chromatin Accessibility Reveals Regulators of Chromatin Accessibility
- PMID: 28118358
- PMCID: PMC5261565
- DOI: 10.1371/journal.pcbi.1005311
Genome-Wide Association between Transcription Factor Expression and Chromatin Accessibility Reveals Regulators of Chromatin Accessibility
Abstract
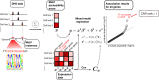
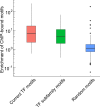
To better understand genome regulation, it is important to uncover the role of transcription factors in the process of chromatin structure establishment and maintenance. Here we present a data-driven approach to systematically characterise transcription factors that are relevant for this process. Our method uses a linear mixed modelling approach to combine datasets of transcription factor binding motif enrichments in open chromatin and gene expression across the same set of cell lines. Applying this approach to the ENCODE dataset, we confirm already known and imply numerous novel transcription factors that play a role in the establishment or maintenance of open chromatin. In particular, our approach rediscovers many factors that have been annotated as pioneer factors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9: 279–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

