DNA Polymerases ImuC and DinB Are Involved in DNA Alkylation Damage Tolerance in Pseudomonas aeruginosa and Pseudomonas putida
- PMID: 28118378
- PMCID: PMC5261740
- DOI: 10.1371/journal.pone.0170719
DNA Polymerases ImuC and DinB Are Involved in DNA Alkylation Damage Tolerance in Pseudomonas aeruginosa and Pseudomonas putida
Abstract
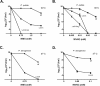
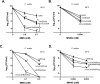
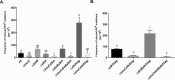
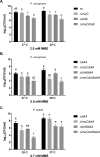
Translesion DNA synthesis (TLS), facilitated by low-fidelity polymerases, is an important DNA damage tolerance mechanism. Here, we investigated the role and biological function of TLS polymerase ImuC (former DnaE2), generally present in bacteria lacking DNA polymerase V, and TLS polymerase DinB in response to DNA alkylation damage in Pseudomonas aeruginosa and P. putida. We found that TLS DNA polymerases ImuC and DinB ensured a protective role against N- and O-methylation induced by N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) in both P. aeruginosa and P. putida. DinB also appeared to be important for the survival of P. aeruginosa and rapidly growing P. putida cells in the presence of methyl methanesulfonate (MMS). The role of ImuC in protection against MMS-induced damage was uncovered under DinB-deficient conditions. Apart from this, both ImuC and DinB were critical for the survival of bacteria with impaired base excision repair (BER) functions upon alkylation damage, lacking DNA glycosylases AlkA and/or Tag. Here, the increased sensitivity of imuCdinB double deficient strains in comparison to single mutants suggested that the specificity of alkylated DNA lesion bypass of DinB and ImuC might also be different. Moreover, our results demonstrated that mutagenesis induced by MMS in pseudomonads was largely ImuC-dependent. Unexpectedly, we discovered that the growth temperature of bacteria affected the efficiency of DinB and ImuC in ensuring cell survival upon alkylation damage. Taken together, the results of our study disclosed the involvement of ImuC in DNA alkylation damage tolerance, especially at low temperatures, and its possible contribution to the adaptation of pseudomonads upon DNA alkylation damage via increased mutagenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






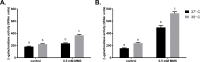
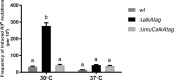

References
-
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231: 11–30. - PubMed
-
- Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150: 77–84. - PubMed
-
- Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, et al. Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair (Amst). 2004;3: 13891407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

