Single-cell epigenomic variability reveals functional cancer heterogeneity
- PMID: 28118844
- PMCID: PMC5259890
- DOI: 10.1186/s13059-016-1133-7
Single-cell epigenomic variability reveals functional cancer heterogeneity
Abstract
Background: Cell-to-cell heterogeneity is a major driver of cancer evolution, progression, and emergence of drug resistance. Epigenomic variation at the single-cell level can rapidly create cancer heterogeneity but is difficult to detect and assess functionally.
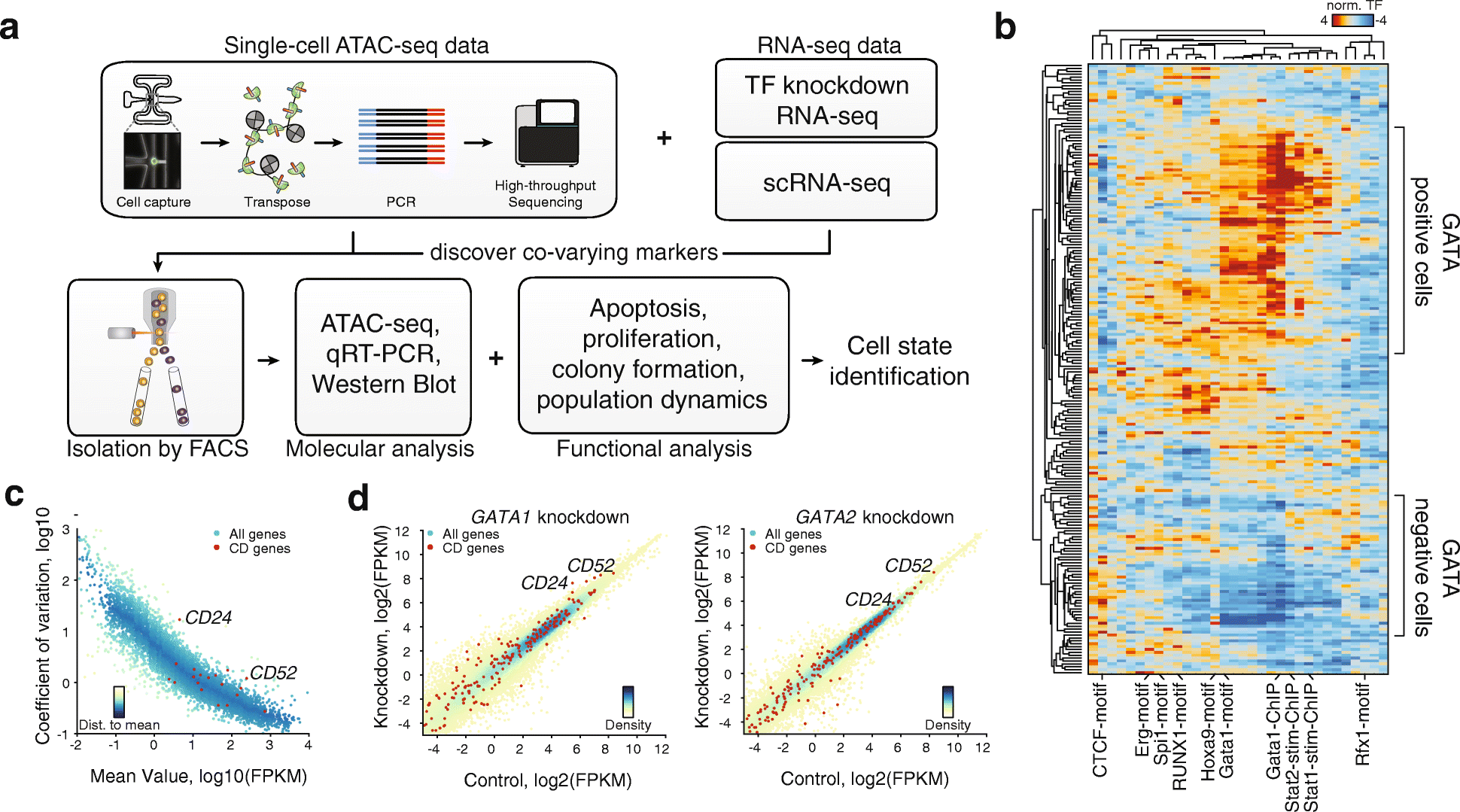
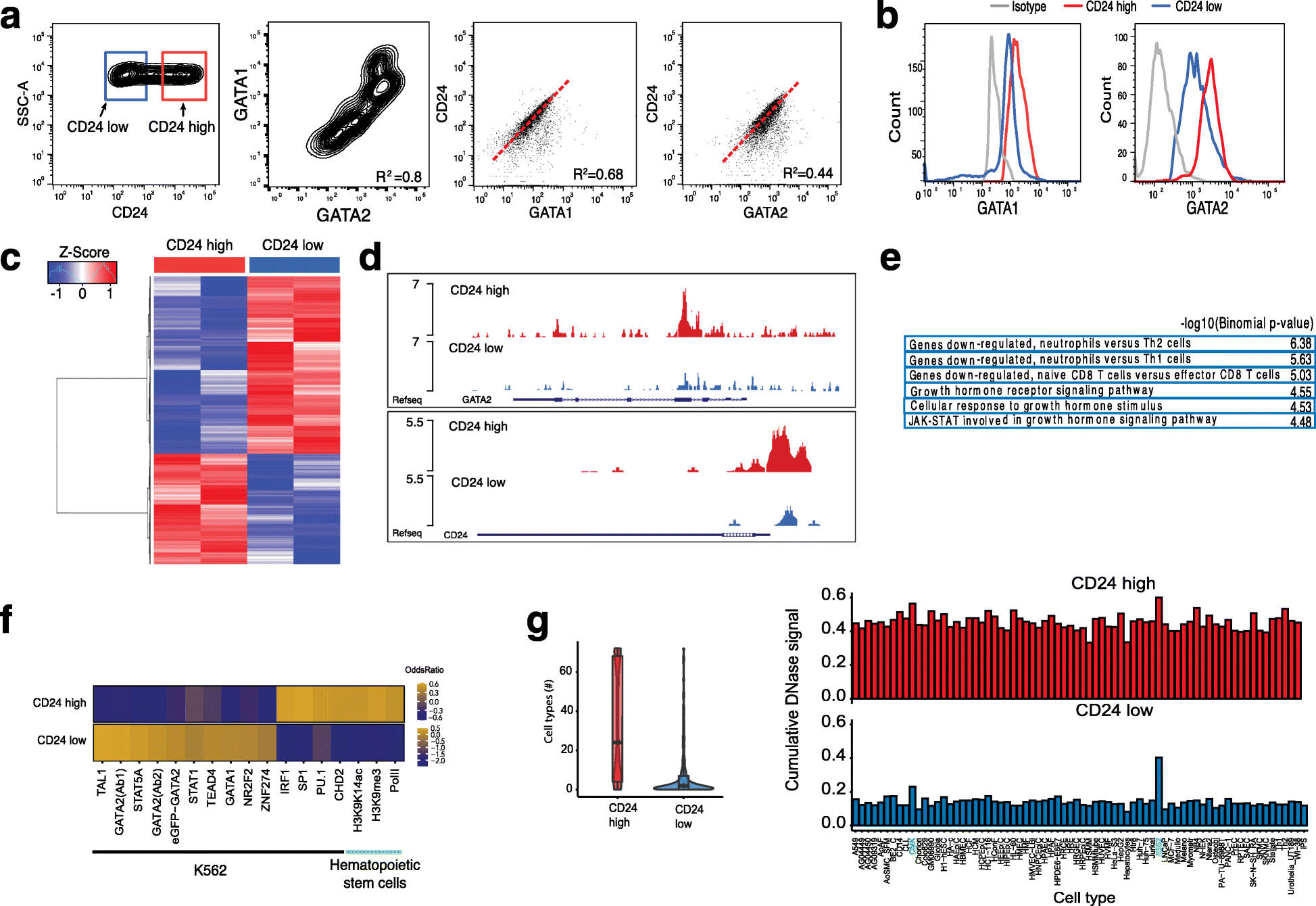
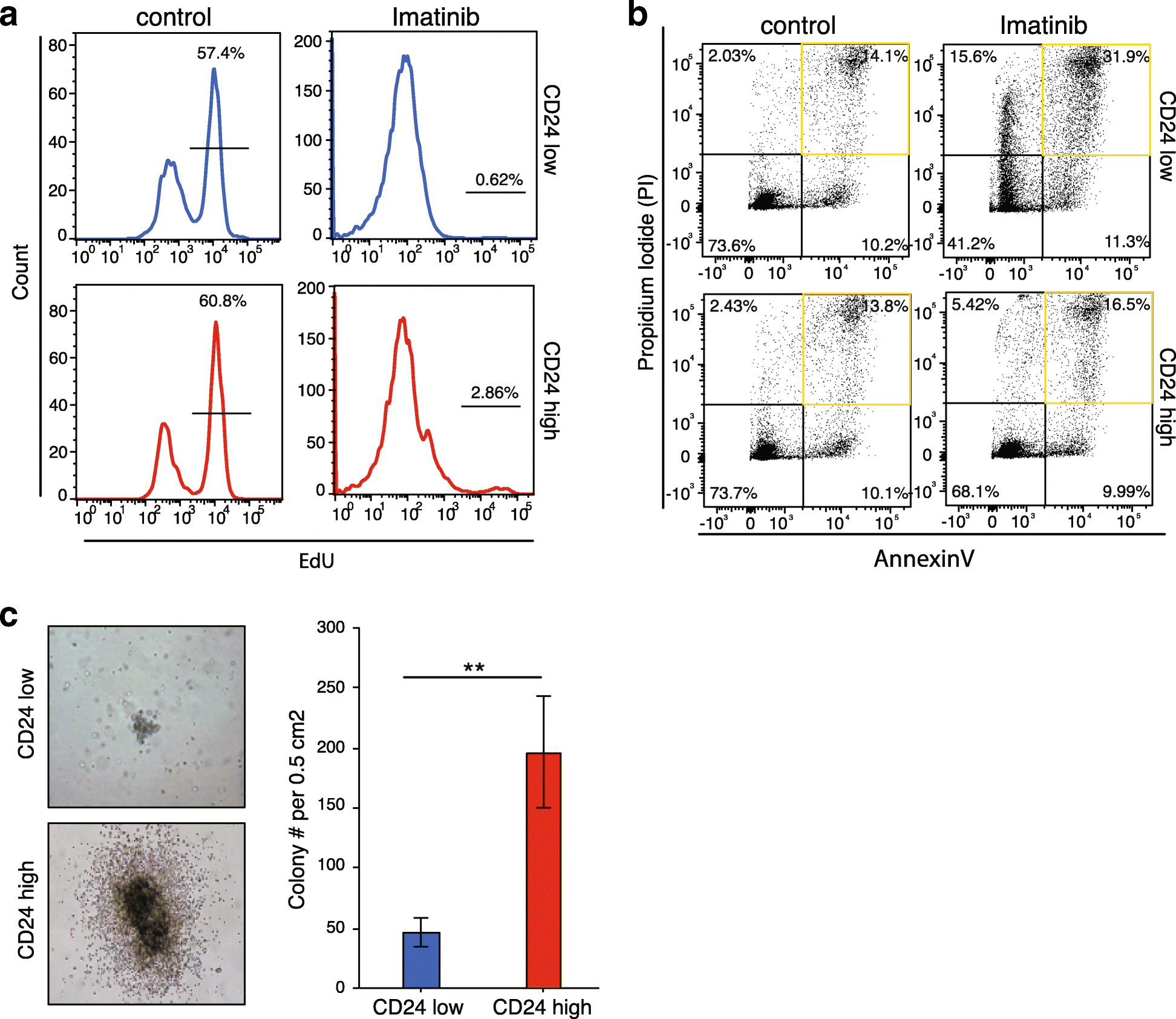
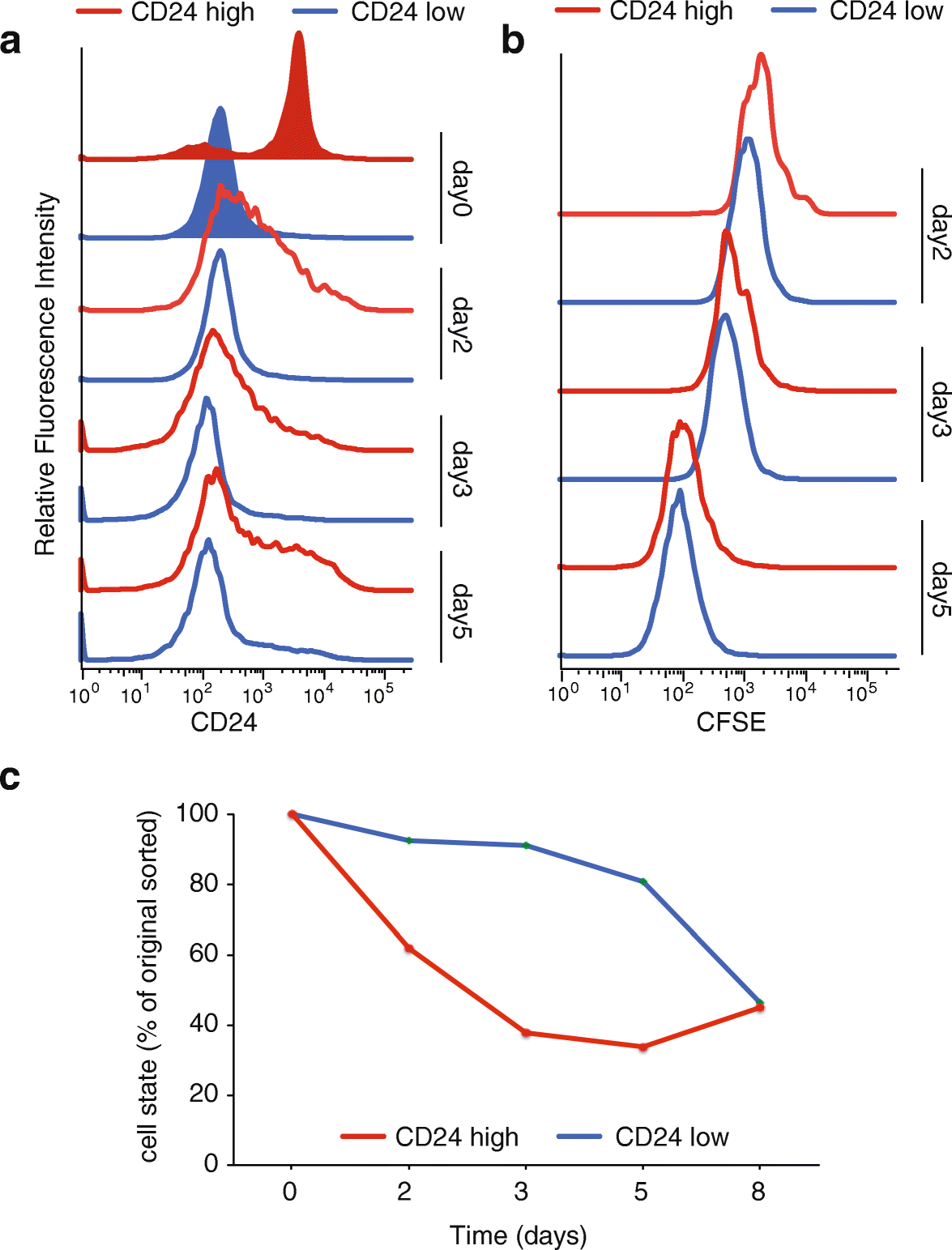
Results: We develop a strategy to bridge the gap between measurement and function in single-cell epigenomics. Using single-cell chromatin accessibility and RNA-seq data in K562 leukemic cells, we identify the cell surface marker CD24 as co-varying with chromatin accessibility changes linked to GATA transcription factors in single cells. Fluorescence-activated cell sorting of CD24 high versus low cells prospectively isolated GATA1 and GATA2 high versus low cells. GATA high versus low cells express differential gene regulatory networks, differential sensitivity to the drug imatinib mesylate, and differential self-renewal capacity. Lineage tracing experiments show that GATA/CD24hi cells have the capability to rapidly reconstitute the heterogeneity within the entire starting population, suggesting that GATA expression levels drive a phenotypically relevant source of epigenomic plasticity.
Conclusion: Single-cell chromatin accessibility can guide prospective characterization of cancer heterogeneity. Epigenomic subpopulations in cancer impact drug sensitivity and the clonal dynamics of cancer evolution.
Keywords: Cancer stem cells; Gene expression noise; Open chromatin.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

