TLR2 2258 G>A single nucleotide polymorphism and the risk of congenital infection with human cytomegalovirus
- PMID: 28118851
- PMCID: PMC5260049
- DOI: 10.1186/s12985-016-0679-z
TLR2 2258 G>A single nucleotide polymorphism and the risk of congenital infection with human cytomegalovirus
Abstract
Background: Human cytomegalovirus (HCMV) is responsible for the most common intrauterine infections, which may be acquired congenitally from infected pregnant woman to fetus. The research was aimed to estimate the role of three single nucleotide polymorphisms (SNPs) located in TLR2 gene, and the common contribution of TLR2, and previously studied TLR4 and TLR9 SNPs, to the occurrence of congenital HCMV infection in fetuses and newborns.
Methods: The study was performed in 20 Polish fetuses and newborns, congenitally infected with HCMV, and in 31 uninfected controls, as well as with participation of pregnant women, the mothers of 16 infected and 14 uninfected offsprings. Genotypes in TLR2 SNPs were determined, using self-designed nested PCR-RFLP assays, and confirmed by sequencing. The genotypes were tested for Hardy-Weinberg (H-W) equilibrium, and for their relationship with the development of congenital cytomegaly, using a logistic regression model. The common influence of TLR2, TLR4 and TLR9 SNPs on the occurrence of congenital disease was estimated by multiple-SNP analysis.
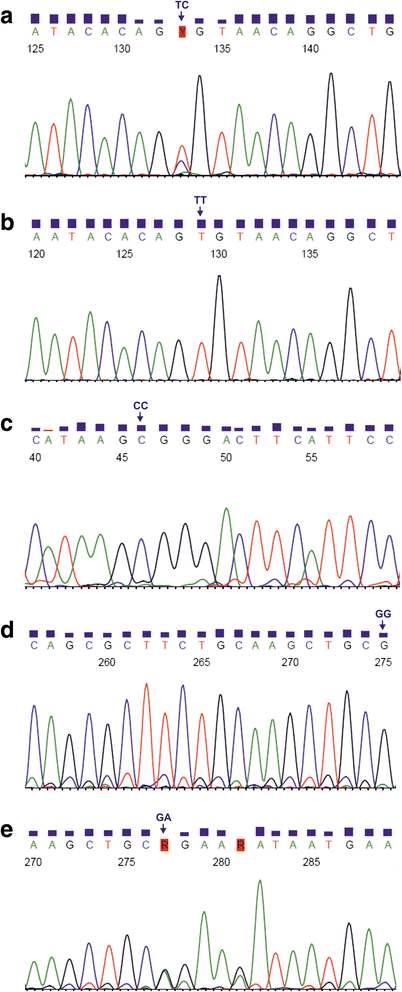
Results: Distribution of the genotypes and alleles in TLR2 1350 T>C and 2029 C>T SNPs was similar between the studied groups of fetuses and neonates. In case of 2258 G>A polymorphism, the GA heterozygotic status was significantly more frequent in the infected cases than among the uninfected individuals (25.0% vs. 3.2%, respectively), and increased the risk of HCMV infection (OR 10.00, 95% CI 1.07-93.44; P ≤ 0.050). Similarly, the A allele within 2258 G>A polymorphism was significantly more frequent among the infected offsprings than in the uninfected ones (12.5% vs. 1.6%; P ≤ 0.050). Complex AA variants for both TLR2 2258 and TLR9 2848 G>A polymorphisms, were estimated to be at increased risk of congenital HCMV infection (OR 11.58, 95% CI 1.19-112.59; P ≤ 0.050). Additionally, significant relationships were observed between the occurrence of complex AA or GA variants for both TLR2 and TLR9 SNPs and the increased viral loads, determined in fetal amniotic fluids and in maternal blood or urine specimens (P ≤ 0.050).
Conclusions: Among various TLR2, TLR4 and TLR9 polymorphisms, TLR2 2258 G>A SNP seems to be an important factor associated with increased risk of congenital HCMV infection in Polish fetuses and neonates.
Keywords: Congenital cytomegaly; Human cytomegalovirus (HCMV); Pregnancy; Single nucleotide polymorphism (SNP); Toll-like receptor 2 (TLR2).
Figures

Similar articles
-
Toll-like receptors genes polymorphisms and the occurrence of HCMV infection among pregnant women.Virol J. 2017 Mar 24;14(1):64. doi: 10.1186/s12985-017-0730-8. Virol J. 2017. PMID: 28340580 Free PMC article.
-
TLR9 2848 GA heterozygotic status possibly predisposes fetuses and newborns to congenital infection with human cytomegalovirus.PLoS One. 2015 Apr 6;10(4):e0122831. doi: 10.1371/journal.pone.0122831. eCollection 2015. PLoS One. 2015. PMID: 25844529 Free PMC article.
-
Association of SNPs from IL1A, IL1B, and IL6 Genes with Human Cytomegalovirus Infection Among Pregnant Women.Viral Immunol. 2017 May;30(4):288-297. doi: 10.1089/vim.2016.0129. Epub 2017 Feb 2. Viral Immunol. 2017. PMID: 28151075
-
Alterations in TLRs as new molecular markers of congenital infections with Human cytomegalovirus?Pathog Dis. 2014 Feb;70(1):3-16. doi: 10.1111/2049-632X.12083. Epub 2013 Sep 10. Pathog Dis. 2014. PMID: 23929630 Review.
-
Maternal Immunity and the Natural History of Congenital Human Cytomegalovirus Infection.Viruses. 2018 Aug 3;10(8):405. doi: 10.3390/v10080405. Viruses. 2018. PMID: 30081449 Free PMC article. Review.
Cited by
-
Toll-like receptors genes polymorphisms and the occurrence of HCMV infection among pregnant women.Virol J. 2017 Mar 24;14(1):64. doi: 10.1186/s12985-017-0730-8. Virol J. 2017. PMID: 28340580 Free PMC article.
-
Pluripotent Stem Cell-Based Models: A Peephole into Virus Infections during Early Pregnancy.Cells. 2020 Feb 26;9(3):542. doi: 10.3390/cells9030542. Cells. 2020. PMID: 32110999 Free PMC article. Review.
-
Single Nucleotide Polymorphisms of Interleukins and Toll-like Receptors and Neuroimaging Results in Newborns with Congenital HCMV Infection.Viruses. 2021 Sep 7;13(9):1783. doi: 10.3390/v13091783. Viruses. 2021. PMID: 34578364 Free PMC article.
-
Association between single nucleotide polymorphisms (SNPs) of IL1, IL12, IL28 and TLR4 and symptoms of congenital cytomegalovirus infection.PLoS One. 2020 May 18;15(5):e0233096. doi: 10.1371/journal.pone.0233096. eCollection 2020. PLoS One. 2020. PMID: 32421725 Free PMC article.
-
Beyond hearing loss: exploring neurological and neurodevelopmental sequelae in asymptomatic congenital cytomegalovirus infection.Pediatr Res. 2025 Jul 8. doi: 10.1038/s41390-025-04232-5. Online ahead of print. Pediatr Res. 2025. PMID: 40629082 Review.
References
-
- Britt W. Cytomegalovirus. In: Remington JS, Klein JO, editors. Infectious diseases of the fetus and newborn infant. 7. Philadelphia: Elsevier Saunders; 2011. pp. 707–56.
-
- Gaj Z, Rycel M, Wilczyński J, Nowakowska D. Seroprevalence of cytomegalovirus infection in the population of Polish pregnant women. Ginekol Pol. 2012;83:337–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

