Large-scale analysis of branchpoint usage across species and cell lines
- PMID: 28119336
- PMCID: PMC5378181
- DOI: 10.1101/gr.202820.115
Large-scale analysis of branchpoint usage across species and cell lines
Abstract
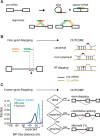
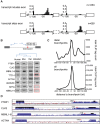
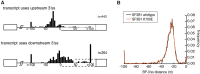
The coding sequence of each human pre-mRNA is interrupted, on average, by 11 introns that must be spliced out for proper gene expression. Each intron contains three obligate signals: a 5' splice site, a branch site, and a 3' splice site. Splice site usage has been mapped exhaustively across different species, cell types, and cellular states. In contrast, only a small fraction of branch sites have been identified even once. The few reported annotations of branch site are imprecise as reverse transcriptase skips several nucleotides while traversing a 2-5 linkage. Here, we report large-scale mapping of the branchpoints from deep sequencing data in three different species and in the SF3B1 K700E oncogenic mutant background. We have developed a novel method whereby raw lariat reads are refined by U2snRNP/pre-mRNA base-pairing models to return the largest current data set of branchpoint sequences with quality metrics. This analysis discovers novel modes of U2snRNA:pre-mRNA base-pairing conserved in yeast and provides insight into the biogenesis of intron circles. Finally, matching branch site usage with isoform selection across the extensive panel of ENCODE RNA-seq data sets offers insight into the mechanisms by which branchpoint usage drives alternative splicing.
© 2017 Taggart et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Bonnal S, Vigevani L, Valcarcel J. 2012. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 11: 847–859. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
