The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases
- PMID: 28119442
- PMCID: PMC5335588
- DOI: 10.1194/jlr.O072629
The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases
Abstract
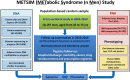
The Metabolic Syndrome in Men (METSIM) study is a population-based study including 10,197 Finnish men examined in 2005-2010. The aim of the study is to investigate nongenetic and genetic factors associated with the risk of T2D and CVD, and with cardiovascular risk factors. The protocol includes a detailed phenotyping of the participants, an oral glucose tolerance test, fasting laboratory measurements including proton NMR measurements, mass spectometry metabolomics, adipose tissue biopsies from 1,400 participants, and a stool sample. In our ongoing follow-up study, we have, to date, reexamined 6,496 participants. Extensive genotyping and exome sequencing have been performed for essentially all METSIM participants, and >2,000 METSIM participants have been whole-genome sequenced. We have identified several nongenetic markers associated with the development of diabetes and cardiovascular events, and participated in several genetic association studies to identify gene variants associated with diabetes, hyperglycemia, and cardiovascular risk factors. The generation of a phenotype and genotype resource in the METSIM study allows us to proceed toward a "systems genetics" approach, which includes statistical methods to quantitate and integrate intermediate phenotypes, such as transcript, protein, or metabolite levels, to provide a global view of the molecular architecture of complex traits.
Keywords: METSIM; cardiovascular risk factors; coronary artery disease; metabolic disease; type 2 diabetes.
Figures
References
-
- Lindström J., and Tuomilehto J.. 2003. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 26: 725–731. - PubMed
-
- Stančáková A., Paananen J., Soininen P., Kangas A. J., Bonnycastle L. L., Morken M. A., Collins F. S., Jackson A. U., Boehnke M. L., Kuusisto J., et al. 2011. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes. 60: 1608–1616. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

