The Arabidopsis thaliana Nuclear Factor Y Transcription Factors
- PMID: 28119722
- PMCID: PMC5222873
- DOI: 10.3389/fpls.2016.02045
The Arabidopsis thaliana Nuclear Factor Y Transcription Factors
Abstract
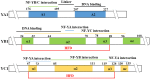
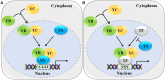
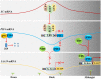
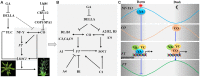
Nuclear factor Y (NF-Y) is an evolutionarily conserved trimeric transcription factor complex present in nearly all eukaryotes. The heterotrimeric NF-Y complex consists of three subunits, NF-YA, NF-YB, and NF-YC, and binds to the CCAAT box in the promoter regions of its target genes to regulate their expression. Yeast and mammal genomes generally have single genes with multiple splicing isoforms that encode each NF-Y subunit. By contrast, plant genomes generally have multi-gene families encoding each subunit and these genes are differentially expressed in various tissues or stages. Therefore, different subunit combinations can lead to a wide variety of NF-Y complexes in various tissues, stages, and growth conditions, indicating the potentially diverse functions of this complex in plants. Indeed, many recent studies have proved that the NF-Y complex plays multiple essential roles in plant growth, development, and stress responses. In this review, we highlight recent progress on NF-Y in Arabidopsis thaliana, including NF-Y protein structure, heterotrimeric complex formation, and the molecular mechanism by which NF-Y regulates downstream target gene expression. We then focus on its biological functions and underlying molecular mechanisms. Finally, possible directions for future research on NF-Y are also presented.
Keywords: Arabidopsis; CCAAT box; abiotic response; flowering time; hypocotyl elongation; nuclear factor Y.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

