Kernel-based Joint Feature Selection and Max-Margin Classification for Early Diagnosis of Parkinson's Disease
- PMID: 28120883
- PMCID: PMC5264393
- DOI: 10.1038/srep41069
Kernel-based Joint Feature Selection and Max-Margin Classification for Early Diagnosis of Parkinson's Disease
Abstract
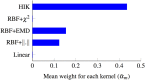
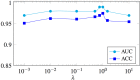
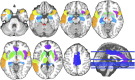
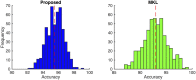
Feature selection methods usually select the most compact and relevant set of features based on their contribution to a linear regression model. Thus, these features might not be the best for a non-linear classifier. This is especially crucial for the tasks, in which the performance is heavily dependent on the feature selection techniques, like the diagnosis of neurodegenerative diseases. Parkinson's disease (PD) is one of the most common neurodegenerative disorders, which progresses slowly while affects the quality of life dramatically. In this paper, we use the data acquired from multi-modal neuroimaging data to diagnose PD by investigating the brain regions, known to be affected at the early stages. We propose a joint kernel-based feature selection and classification framework. Unlike conventional feature selection techniques that select features based on their performance in the original input feature space, we select features that best benefit the classification scheme in the kernel space. We further propose kernel functions, specifically designed for our non-negative feature types. We use MRI and SPECT data of 538 subjects from the PPMI database, and obtain a diagnosis accuracy of 97.5%, which outperforms all baseline and state-of-the-art methods.
Figures
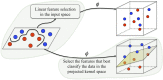
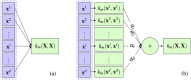
 denotes a non-linear mapping function.
denotes a non-linear mapping function.

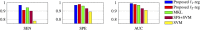

References
-
- Adeli-Mosabbeb E., Thung K.-H., An L., Shi F. & Shen D. Robust feature-sample linear discriminant analysis for brain disorders diagnosis. In NIPS (2015).
-
- Gu Q., Li Z. & Han J. Generalized fisher score for feature selection. In UAI (2011).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

