Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change
- PMID: 28121018
- PMCID: PMC5382103
- DOI: 10.1111/evo.13193
Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change
Abstract
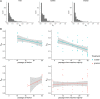
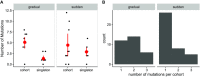
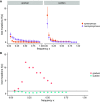
Understanding the dynamics of molecular adaptation is a fundamental goal of evolutionary biology. While adaptation to constant environments has been well characterized, the effects of environmental complexity remain seldom studied. One simple but understudied factor is the rate of environmental change. Here we used experimental evolution with RNA viruses to investigate whether evolutionary dynamics varied based on the rate of environmental turnover. We used whole-genome next-generation sequencing to characterize evolutionary dynamics in virus populations adapting to a sudden versus gradual shift onto a novel host cell type. In support of theoretical models, we found that when populations evolved in response to a sudden environmental change, mutations of large beneficial effect tended to fix early, followed by mutations of smaller beneficial effect; as predicted, this pattern broke down in response to a gradual environmental change. Early mutational steps were highly parallel across replicate populations in both treatments. The fixation of single mutations was less common than sweeps of associated "cohorts" of mutations, and this pattern intensified when the environment changed gradually. Additionally, clonal interference appeared stronger in response to a gradual change. Our results suggest that the rate of environmental change is an important determinant of evolutionary dynamics in asexual populations.
Keywords: Adaptation; Sindbis virus; clonal interference; epistasis; experimental evolution.
© 2017 The Author(s). Evolution published by Wiley Periodicals, Inc. on behalf of The Society for the Study of Evolution.
Figures

Similar articles
-
Rate of novel host invasion affects adaptability of evolving RNA virus lineages.Proc Biol Sci. 2015 Aug 22;282(1813):20150801. doi: 10.1098/rspb.2015.0801. Proc Biol Sci. 2015. PMID: 26246544 Free PMC article.
-
Genomics of Adaptation Depends on the Rate of Environmental Change in Experimental Yeast Populations.Mol Biol Evol. 2017 Oct 1;34(10):2613-2626. doi: 10.1093/molbev/msx185. Mol Biol Evol. 2017. PMID: 28957501
-
Experimental evolution and genome sequencing reveal variation in levels of clonal interference in large populations of bacteriophage phiX174.BMC Evol Biol. 2008 Mar 17;8:85. doi: 10.1186/1471-2148-8-85. BMC Evol Biol. 2008. PMID: 18366653 Free PMC article.
-
The spectrum of adaptive mutations in experimental evolution.Genomics. 2014 Dec;104(6 Pt A):412-6. doi: 10.1016/j.ygeno.2014.09.011. Epub 2014 Sep 28. Genomics. 2014. PMID: 25269377 Free PMC article. Review.
-
Fitness effects of beneficial mutations: the mutational landscape model in experimental evolution.Curr Opin Genet Dev. 2006 Dec;16(6):618-23. doi: 10.1016/j.gde.2006.10.006. Epub 2006 Oct 19. Curr Opin Genet Dev. 2006. PMID: 17055718 Review.
Cited by
-
Profile of Paul E. Turner.Proc Natl Acad Sci U S A. 2020 Aug 11;117(32):18906-18908. doi: 10.1073/pnas.2014283117. Epub 2020 Aug 3. Proc Natl Acad Sci U S A. 2020. PMID: 32747551 Free PMC article. No abstract available.
-
A network-based approach to deciphering a dynamic microbiome's response to a subtle perturbation.Sci Rep. 2020 Nov 11;10(1):19530. doi: 10.1038/s41598-020-73920-5. Sci Rep. 2020. PMID: 33177547 Free PMC article.
-
Fluctuating selection facilitates the discovery of broadly effective but difficult to reach adaptive outcomes in yeast.Evol Lett. 2023 Nov 10;8(2):243-252. doi: 10.1093/evlett/qrad055. eCollection 2024 Apr. Evol Lett. 2023. PMID: 38525031 Free PMC article.
-
Intra-Population Competition during Adaptation to Increased Temperature in an RNA Bacteriophage.Int J Mol Sci. 2021 Jun 24;22(13):6815. doi: 10.3390/ijms22136815. Int J Mol Sci. 2021. PMID: 34202838 Free PMC article.
-
Fluctuating Environments Maintain Genetic Diversity through Neutral Fitness Effects and Balancing Selection.Mol Biol Evol. 2021 Sep 27;38(10):4362-4375. doi: 10.1093/molbev/msab173. Mol Biol Evol. 2021. PMID: 34132791 Free PMC article.
References
-
- Bello, Y. , and Waxman D.. 2006. Near‐periodic substitution and the genetic variance induced by environmental change. J. Theor. Biol. 239:152–160. - PubMed
-
- Berger, E. A. , Murphy P. M., and Farber J. M.. 1999. Chemokine receptors as HIV‐1 coreceptors: roles in viral entry, tropism, and disease. Ann. Rev. Immunol. 17:657–700. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
