Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq
- PMID: 28121289
- PMCID: PMC5295818
- DOI: 10.7554/eLife.22039
Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq
Abstract
Understanding the mechanisms that regulate cell type-specific transcriptional programs requires developing a lexicon of their genomic regulatory elements. We developed a lineage-selective method to map transcriptional enhancers, regulatory genomic regions that activate transcription, in mice. Since most tissue-specific enhancers are bound by the transcriptional co-activator Ep300, we used Cre-directed, lineage-specific Ep300 biotinylation and pulldown on immobilized streptavidin followed by next generation sequencing of co-precipitated DNA to identify lineage-specific enhancers. By driving this system with lineage-specific Cre transgenes, we mapped enhancers active in embryonic endothelial cells/blood or skeletal muscle. Analysis of these enhancers identified new transcription factor heterodimer motifs that likely regulate transcription in these lineages. Furthermore, we identified candidate enhancers that regulate adult heart- or lung- specific endothelial cell specialization. Our strategy for tissue-specific protein biotinylation opens new avenues for studying lineage-specific protein-DNA and protein-protein interactions.
Keywords: angiogenesis; chromosomes; developmental biology; endothelial cell; genes; mouse; p300; stem cells; transcriptional enhancer.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






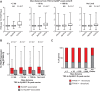






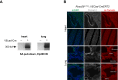


References
-
- Becker PW, Sacilotto N, Nornes S, Neal A, Thomas MO, Liu K, Preece C, Ratnayaka I, Davies B, Bou-Gharios G, De Val S. An intronic Flk1 enhancer directs Arterial-Specific expression via RBPJ-mediated venous repression. Arteriosclerosis, Thrombosis, and Vascular Biology. 2016;36:1209–1219. doi: 10.1161/ATVBAHA.116.307517. - DOI - PMC - PubMed
-
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nature Genetics. 2010;42:806–810. doi: 10.1038/ng.650. - DOI - PMC - PubMed
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genetics. 2012;44:148–156. doi: 10.1038/ng.1064. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

