Refactoring the Embden-Meyerhof-Parnas Pathway as a Whole of Portable GlucoBricks for Implantation of Glycolytic Modules in Gram-Negative Bacteria
- PMID: 28121421
- PMCID: PMC5440799
- DOI: 10.1021/acssynbio.6b00230
Refactoring the Embden-Meyerhof-Parnas Pathway as a Whole of Portable GlucoBricks for Implantation of Glycolytic Modules in Gram-Negative Bacteria
Abstract
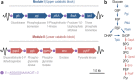
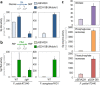
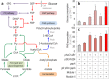
The Embden-Meyerhof-Parnas (EMP) pathway is generally considered to be the biochemical standard for glucose catabolism. Alas, its native genomic organization and the control of gene expression in Escherichia coli are both very intricate, which limits the portability of the EMP pathway to other biotechnologically important bacterial hosts that lack the route. In this work, the genes encoding all the enzymes of the linear EMP route have been individually recruited from the genome of E. coli K-12, edited in silico to remove their endogenous regulatory signals, and synthesized de novo following a standard (GlucoBrick) that enables their grouping in the form of functional modules at the user's will. After verifying their activity in several glycolytic mutants of E. coli, the versatility of these GlucoBricks was demonstrated in quantitative physiology tests and biochemical assays carried out in Pseudomonas putida KT2440 and P. aeruginosa PAO1 as the heterologous hosts. Specific configurations of GlucoBricks were also adopted to streamline the downward circulation of carbon from hexoses to pyruvate in E. coli recombinants, thereby resulting in a 3-fold increase of poly(3-hydroxybutyrate) synthesis from glucose. Refactoring whole metabolic blocks in the fashion described in this work thus eases the engineering of biochemical processes where the optimization of carbon traffic is facilitated by the operation of the EMP pathway-which yields more ATP than other glycolytic routes such as the Entner-Doudoroff pathway.
Keywords: Escherichia coli; PHB; Pseudomonas putida; glycolysis; metabolic engineering; standardization.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Re-Factoring Glycolytic Genes for Targeted Engineering of Catabolism in Gram-Negative Bacteria.Methods Mol Biol. 2018;1772:3-24. doi: 10.1007/978-1-4939-7795-6_1. Methods Mol Biol. 2018. PMID: 29754220
-
Functional implementation of a linear glycolysis for sugar catabolism in Pseudomonas putida.Metab Eng. 2019 Jul;54:200-211. doi: 10.1016/j.ymben.2019.04.005. Epub 2019 Apr 19. Metab Eng. 2019. PMID: 31009747
-
Pseudomonas putida KT2440 Strain Metabolizes Glucose through a Cycle Formed by Enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and Pentose Phosphate Pathways.J Biol Chem. 2015 Oct 23;290(43):25920-32. doi: 10.1074/jbc.M115.687749. Epub 2015 Sep 8. J Biol Chem. 2015. PMID: 26350459 Free PMC article.
-
The Entner-Doudoroff pathway: history, physiology and molecular biology.FEMS Microbiol Rev. 1992 Sep;9(1):1-27. doi: 10.1111/j.1574-6968.1992.tb05822.x. FEMS Microbiol Rev. 1992. PMID: 1389313 Review.
-
Alternative carbohydrate pathways - enzymes, functions and engineering.Crit Rev Biotechnol. 2020 Nov;40(7):895-912. doi: 10.1080/07388551.2020.1785386. Epub 2020 Jul 13. Crit Rev Biotechnol. 2020. PMID: 32654530 Review.
Cited by
-
A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida.Nat Commun. 2020 Oct 7;11(1):5045. doi: 10.1038/s41467-020-18813-x. Nat Commun. 2020. PMID: 33028813 Free PMC article.
-
Past, Present, and Future of Genome Modification in Escherichia coli.Microorganisms. 2022 Sep 14;10(9):1835. doi: 10.3390/microorganisms10091835. Microorganisms. 2022. PMID: 36144436 Free PMC article. Review.
-
Gross transcriptomic analysis of Pseudomonas putida for diagnosing environmental shifts.Microb Biotechnol. 2020 Jan;13(1):263-273. doi: 10.1111/1751-7915.13404. Epub 2019 Apr 7. Microb Biotechnol. 2020. PMID: 30957409 Free PMC article.
-
Synthetic metabolism for in vitro acetone biosynthesis driven by ATP regeneration.RSC Chem Biol. 2022 Sep 16;3(11):1331-1341. doi: 10.1039/d2cb00170e. eCollection 2022 Nov 2. RSC Chem Biol. 2022. PMID: 36349222 Free PMC article.
-
Deciphering the Metabolic Pathway Difference Between Saccharopolyspora pogona and Saccharopolyspora spinosa by Comparative Proteomics and Metabonomics.Front Microbiol. 2020 Mar 18;11:396. doi: 10.3389/fmicb.2020.00396. eCollection 2020. Front Microbiol. 2020. PMID: 32256469 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

