CyNetSVM: A Cytoscape App for Cancer Biomarker Identification Using Network Constrained Support Vector Machines
- PMID: 28122019
- PMCID: PMC5266326
- DOI: 10.1371/journal.pone.0170482
CyNetSVM: A Cytoscape App for Cancer Biomarker Identification Using Network Constrained Support Vector Machines
Abstract
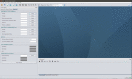
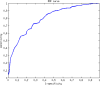
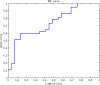
One of the important tasks in cancer research is to identify biomarkers and build classification models for clinical outcome prediction. In this paper, we develop a CyNetSVM software package, implemented in Java and integrated with Cytoscape as an app, to identify network biomarkers using network-constrained support vector machines (NetSVM). The Cytoscape app of NetSVM is specifically designed to improve the usability of NetSVM with the following enhancements: (1) user-friendly graphical user interface (GUI), (2) computationally efficient core program and (3) convenient network visualization capability. The CyNetSVM app has been used to analyze breast cancer data to identify network genes associated with breast cancer recurrence. The biological function of these network genes is enriched in signaling pathways associated with breast cancer progression, showing the effectiveness of CyNetSVM for cancer biomarker identification. The CyNetSVM package is available at Cytoscape App Store and http://sourceforge.net/projects/netsvmjava; a sample data set is also provided at sourceforge.net.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Identifying cancer biomarkers by network-constrained support vector machines.BMC Syst Biol. 2011 Oct 12;5:161. doi: 10.1186/1752-0509-5-161. BMC Syst Biol. 2011. PMID: 21992556 Free PMC article.
-
C-Biomarker.net: A Cytoscape app for the identification of cancer biomarker genes from cores of large biomolecular networks.Biosystems. 2023 Apr;226:104887. doi: 10.1016/j.biosystems.2023.104887. Epub 2023 Mar 28. Biosystems. 2023. PMID: 36990379
-
Development and use of a Cytoscape app for GRNCOP2.Comput Methods Programs Biomed. 2019 Aug;177:211-218. doi: 10.1016/j.cmpb.2019.05.030. Epub 2019 Jun 4. Comput Methods Programs Biomed. 2019. PMID: 31319950
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
Reviewing ensemble classification methods in breast cancer.Comput Methods Programs Biomed. 2019 Aug;177:89-112. doi: 10.1016/j.cmpb.2019.05.019. Epub 2019 May 20. Comput Methods Programs Biomed. 2019. PMID: 31319964 Review.
Cited by
-
Leveraging User-Friendly Network Approaches to Extract Knowledge From High-Throughput Omics Datasets.Front Genet. 2019 Nov 13;10:1120. doi: 10.3389/fgene.2019.01120. eCollection 2019. Front Genet. 2019. PMID: 31798629 Free PMC article. Review.
-
Edgetic perturbation signatures represent known and novel cancer biomarkers.Sci Rep. 2020 Mar 9;10(1):4350. doi: 10.1038/s41598-020-61422-3. Sci Rep. 2020. PMID: 32152446 Free PMC article.
-
Optimizing miRNA-module diagnostic biomarkers of gastric carcinoma via integrated network analysis.PLoS One. 2018 Jun 7;13(6):e0198445. doi: 10.1371/journal.pone.0198445. eCollection 2018. PLoS One. 2018. PMID: 29879180 Free PMC article.
References
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. PubMed Central PMCID: PMC1239896. 10.1073/pnas.0506580102 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

