Germ Cell-less Promotes Centrosome Segregation to Induce Germ Cell Formation
- PMID: 28122234
- PMCID: PMC5327791
- DOI: 10.1016/j.celrep.2016.12.074
Germ Cell-less Promotes Centrosome Segregation to Induce Germ Cell Formation
Abstract
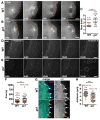
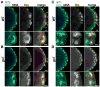
The primordial germ cells (PGCs) specified during embryogenesis serve as progenitors to the adult germline stem cells. In Drosophila, the proper specification and formation of PGCs require both centrosomes and germ plasm, which contains the germline determinants. Centrosomes are microtubule (MT)-organizing centers that ensure the faithful segregation of germ plasm into PGCs. To date, mechanisms that modulate centrosome behavior to engineer PGC development have remained elusive. Only one germ plasm component, Germ cell-less (Gcl), is known to play a role in PGC formation. Here, we show that Gcl engineers PGC formation by regulating centrosome dynamics. Loss of gcl leads to aberrant centrosome separation and elaboration of the astral MT network, resulting in inefficient germ plasm segregation and aborted PGC cellularization. Importantly, compromising centrosome separation alone is sufficient to mimic the gcl loss-of-function phenotypes. We conclude Gcl functions as a key regulator of centrosome separation required for proper PGC development.
Keywords: cell fate; centrosome; germ cell-less; germ cells; germ plasm; stem cells.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- EWEN-CAMPEN B, SCHWAGER EE, EXTAVOUR CG. The molecular machinery of germ line specification. Mol Reprod Dev. 2010;77:3–18. - PubMed
-
- FIELD CM, COUGHLIN M, DOBERSTEIN S, MARTY T, SULLIVAN W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development. 2005;132:2849–2860. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

