Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies
- PMID: 28122605
- PMCID: PMC5267453
- DOI: 10.1186/s13059-016-1143-5
Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies
Abstract
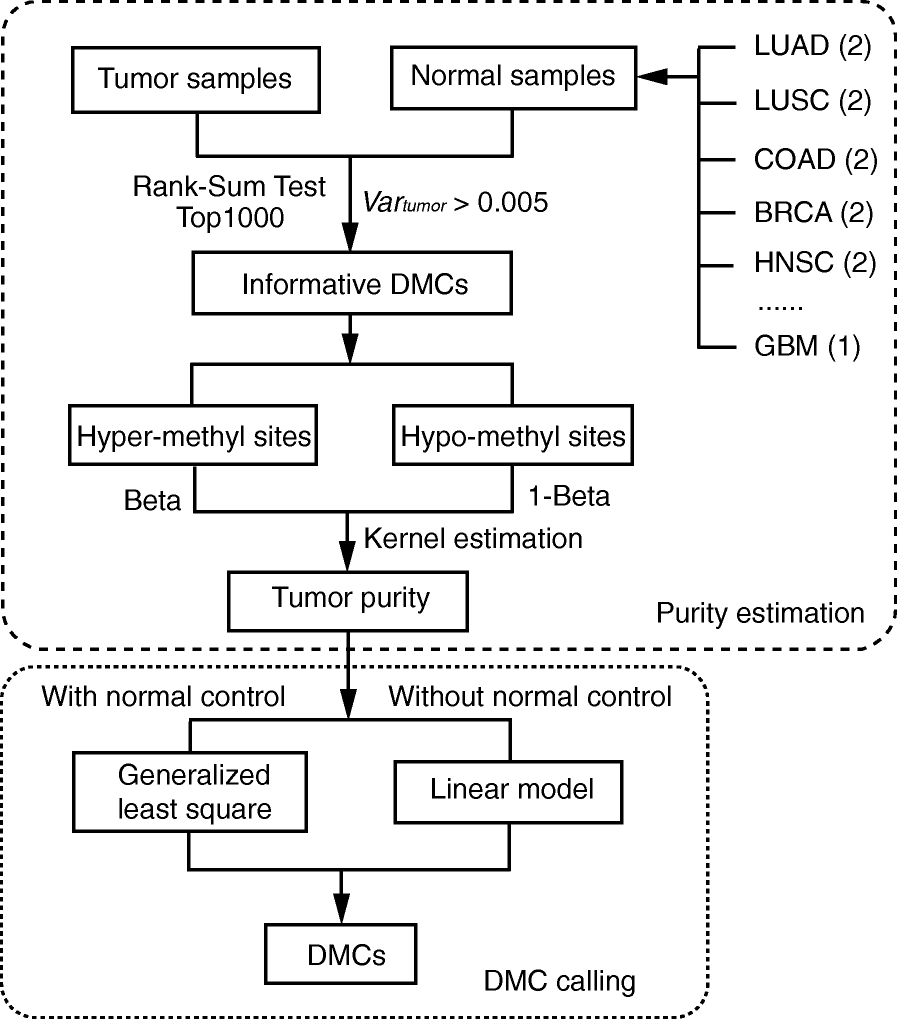
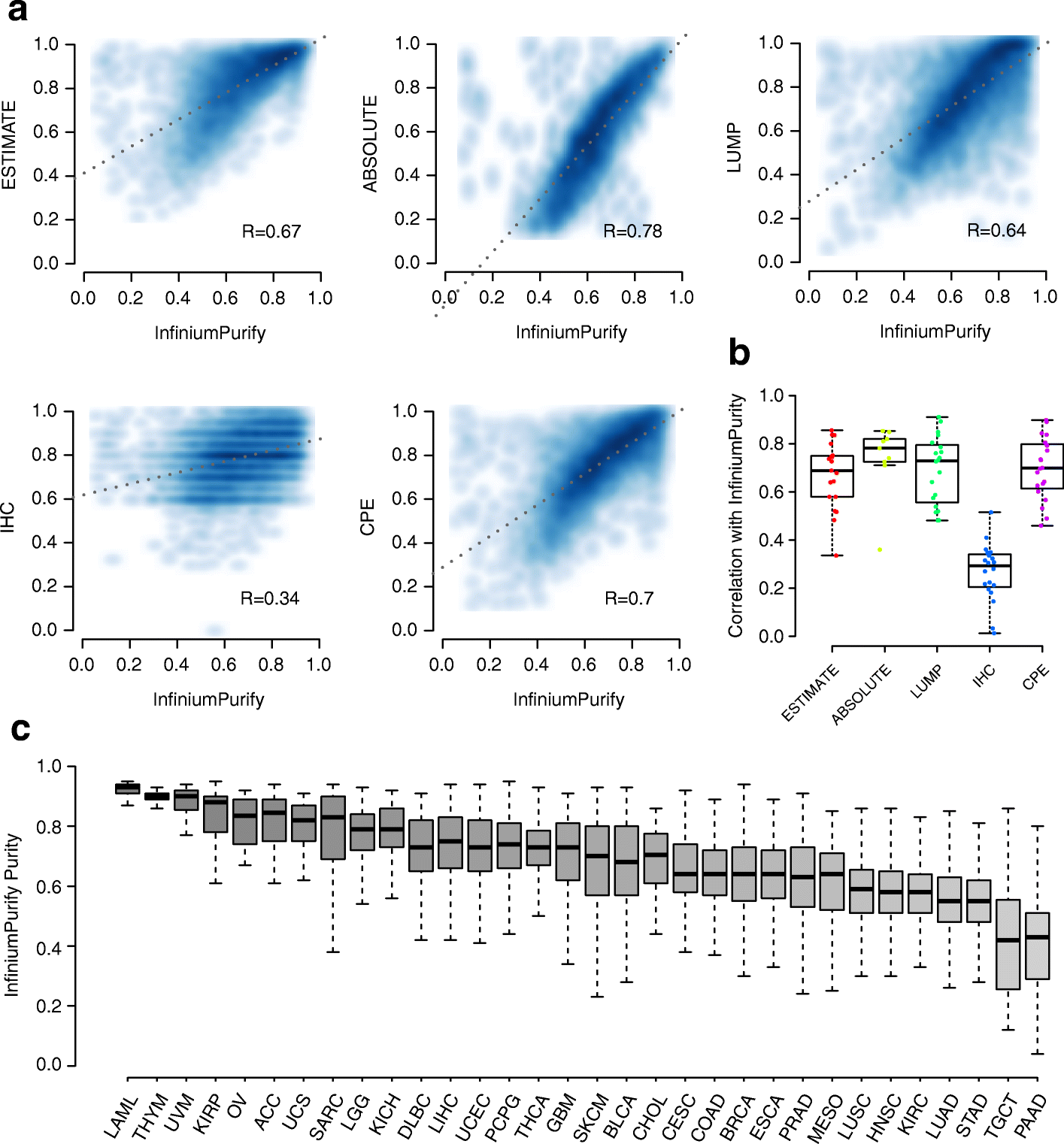
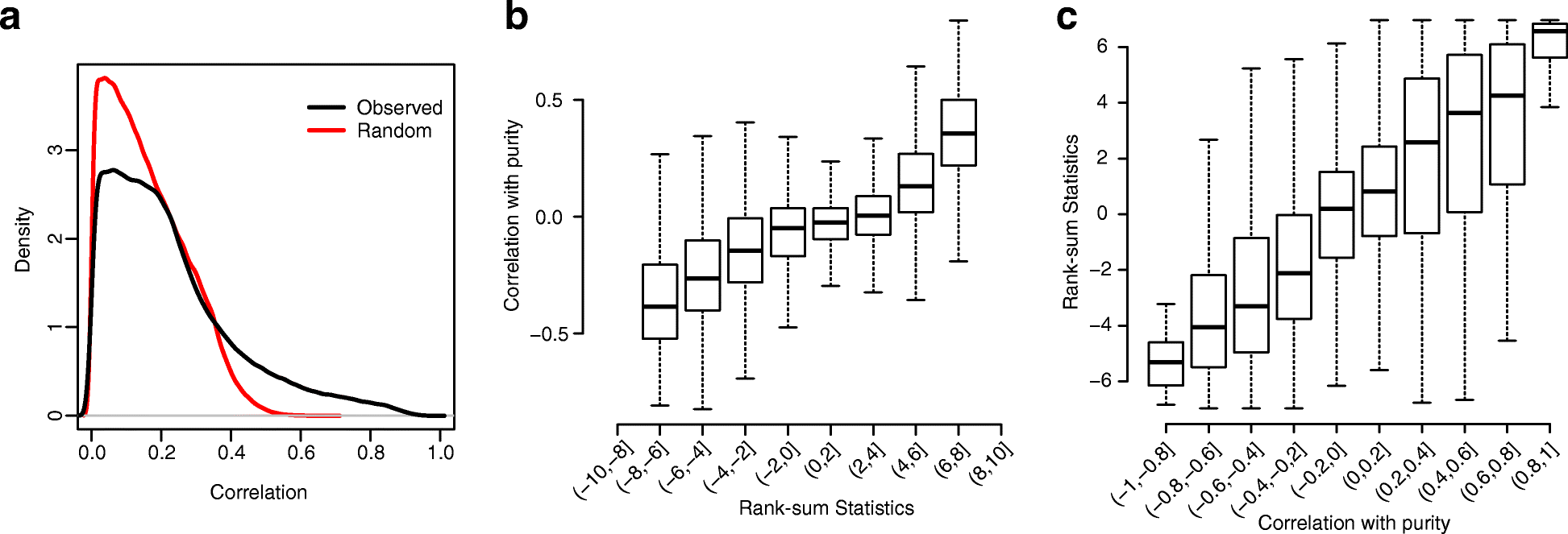
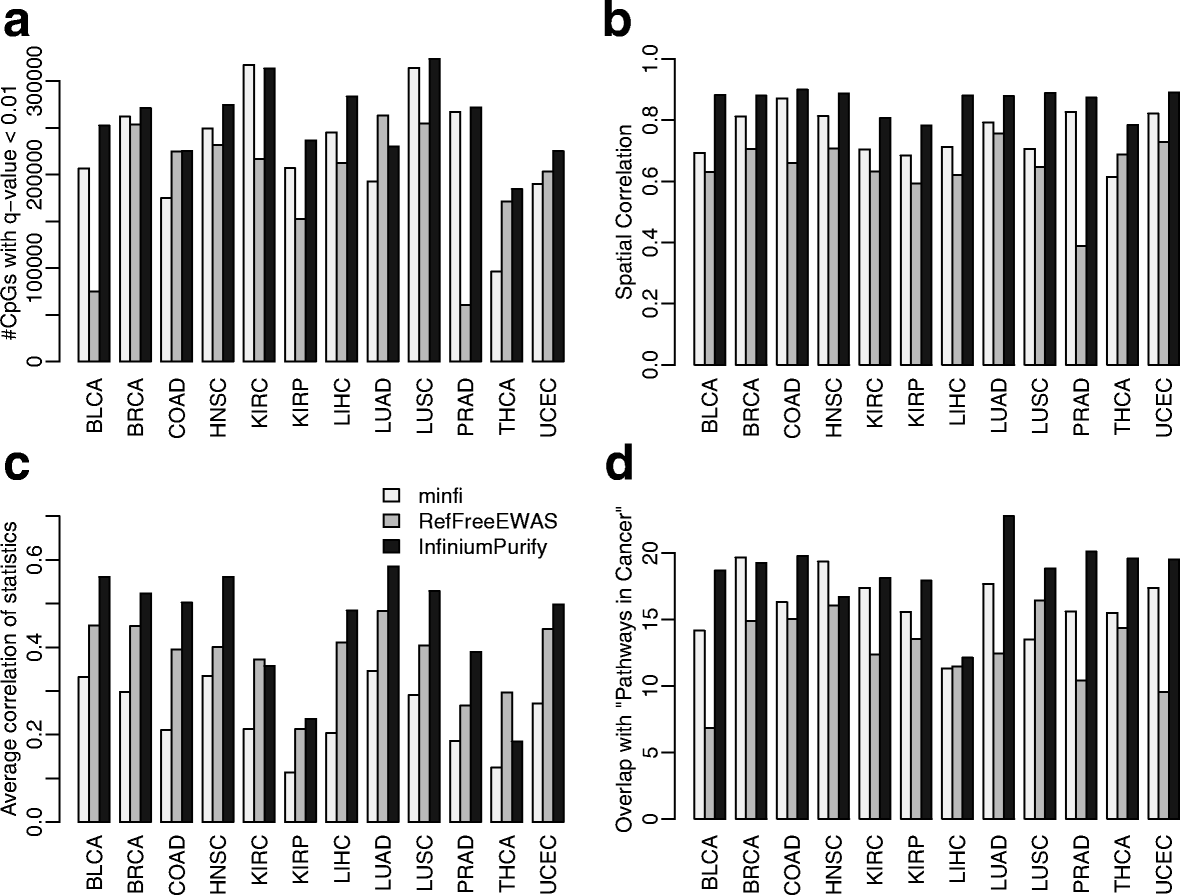
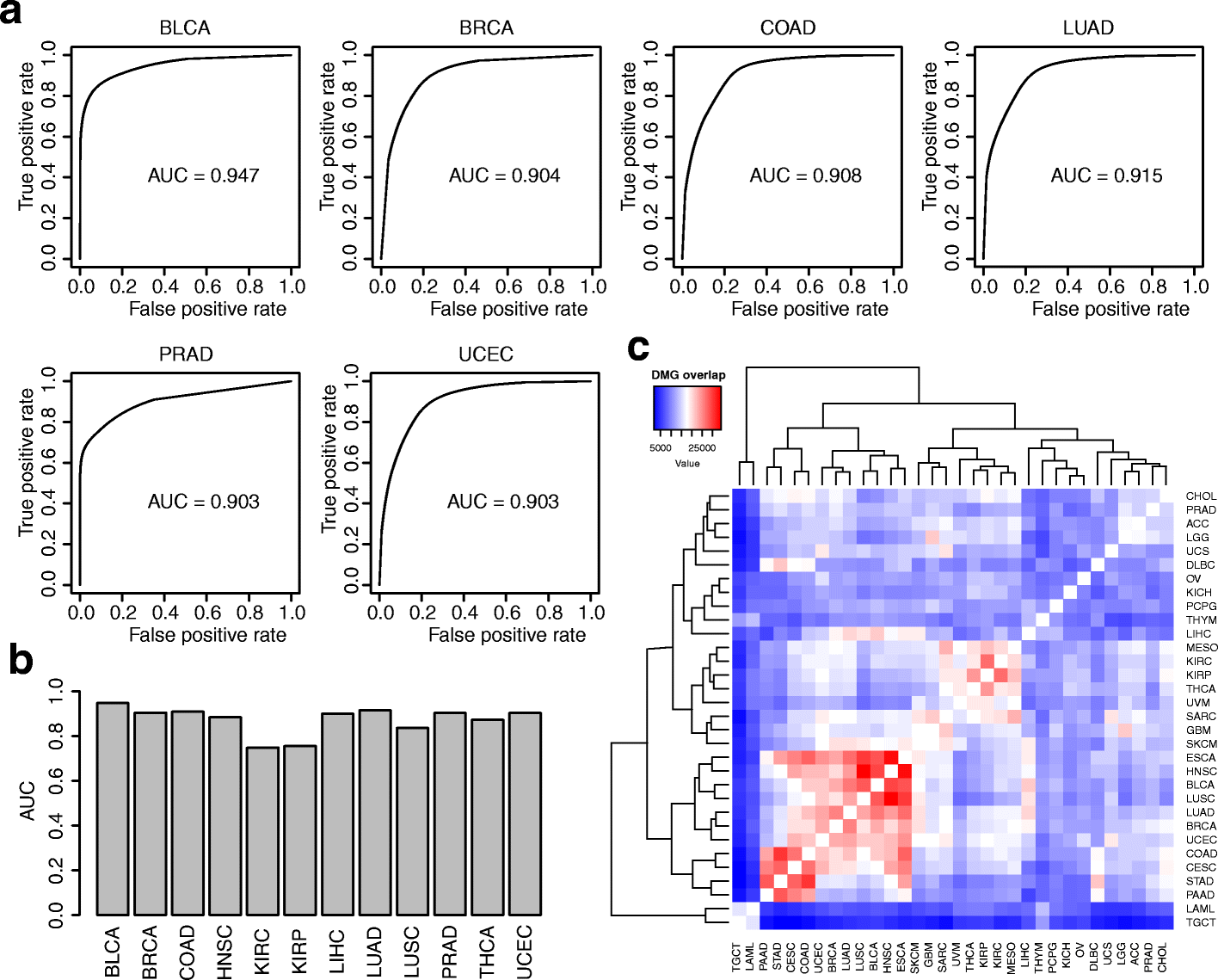
We present a set of statistical methods for the analysis of DNA methylation microarray data, which account for tumor purity. These methods are an extension of our previously developed method for purity estimation; our updated method is flexible, efficient, and does not require data from reference samples or matched normal controls. We also present a method for incorporating purity information for differential methylation analysis. In addition, we propose a control-free differential methylation calling method when normal controls are not available. Extensive analyses of TCGA data demonstrate that our methods provide accurate results. All methods are implemented in InfiniumPurify.
Keywords: Cancer epigenomics; DNA methylation; Differential methylation; Tumor purity.
Figures
Similar articles
-
Tumor purity and differential methylation in cancer epigenomics.Brief Funct Genomics. 2016 Nov;15(6):408-419. doi: 10.1093/bfgp/elw016. Epub 2016 May 19. Brief Funct Genomics. 2016. PMID: 27199459 Review.
-
Universal informative CpG sites for inferring tumor purity from DNA methylation microarray data.J Bioinform Comput Biol. 2018 Jun;16(3):1750030. doi: 10.1142/S0219720017500305. Epub 2017 Dec 28. J Bioinform Comput Biol. 2018. PMID: 29347875
-
CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA.Genome Biol. 2017 Mar 24;18(1):53. doi: 10.1186/s13059-017-1191-5. Genome Biol. 2017. PMID: 28335812 Free PMC article.
-
PEIS: a novel approach of tumor purity estimation by identifying information sites through integrating signal based on DNA methylation data.BMC Bioinformatics. 2019 Dec 30;20(Suppl 22):714. doi: 10.1186/s12859-019-3227-1. BMC Bioinformatics. 2019. PMID: 31888435 Free PMC article.
-
Strategies for validation and testing of DNA methylation biomarkers.Epigenomics. 2014;6(6):603-22. doi: 10.2217/epi.14.43. Epigenomics. 2014. PMID: 25531255 Review.
Cited by
-
High expression of TREM2 promotes EMT via the PI3K/AKT pathway in gastric cancer: bioinformatics analysis and experimental verification.J Cancer. 2021 Apr 2;12(11):3277-3290. doi: 10.7150/jca.55077. eCollection 2021. J Cancer. 2021. PMID: 33976737 Free PMC article.
-
HiTIMED: hierarchical tumor immune microenvironment epigenetic deconvolution for accurate cell type resolution in the tumor microenvironment using tumor-type-specific DNA methylation data.J Transl Med. 2022 Nov 8;20(1):516. doi: 10.1186/s12967-022-03736-6. J Transl Med. 2022. PMID: 36348337 Free PMC article.
-
InfiniumPurify: An R package for estimating and accounting for tumor purity in cancer methylation research.Genes Dis. 2018 Feb 21;5(1):43-45. doi: 10.1016/j.gendis.2018.02.003. eCollection 2018 Mar. Genes Dis. 2018. PMID: 30258934 Free PMC article.
-
A Novel Prognostic Model of Early-Stage Lung Adenocarcinoma Integrating Methylation and Immune Biomarkers.Front Genet. 2021 Jan 21;11:634634. doi: 10.3389/fgene.2020.634634. eCollection 2020. Front Genet. 2021. PMID: 33552145 Free PMC article.
-
Systematic Assessment of Tumor Purity and Its Clinical Implications.JCO Precis Oncol. 2020 Sep 4;4:PO.20.00016. doi: 10.1200/PO.20.00016. eCollection 2020. JCO Precis Oncol. 2020. PMID: 33015524 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

