Identification of potential saliva and tear biomarkers in primary Sjögren's syndrome, utilising the extraction of extracellular vesicles and proteomics analysis
- PMID: 28122643
- PMCID: PMC5264463
- DOI: 10.1186/s13075-017-1228-x
Identification of potential saliva and tear biomarkers in primary Sjögren's syndrome, utilising the extraction of extracellular vesicles and proteomics analysis
Abstract
Background: There is a long-lasting need for non-invasive, more accurate diagnostic techniques when evaluating primary Sjögren's syndrome (pSS) patients. Incorporation of additional diagnostics involving screening for disease-specific biomarkers in biological fluid is a promising concept that requires further investigation. In the current study we aimed to explore novel disease biomarkers in saliva and tears from pSS patients.
Methods: Liquid chromatography-mass spectrometry (LC-MS) was performed on stimulated whole saliva and tears from 27 pSS patients and 32 healthy controls, and salivary and tear proteomic biomarker profiles were generated. LC-MS was also combined with size exclusion chromatography to isolate extracellular vesicles (EVs) from both fluids. Nanoparticle tracking analysis was conducted on joint fractions from the saliva and tears to determine size distribution and concentration of EVs. Further EV characterisation was performed by immunoaffinity capture of CD9-positive EVs using magnetic beads, detected by flow cytometry. The LC-MS data were analysed for quantitative differences between patient and control groups using Scaffold, and the proteins were further analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID), for gene ontology overrepresentation, and the Search Tool for the Retrieval of Interacting Genes/Proteins for protein-protein interaction network analysis.
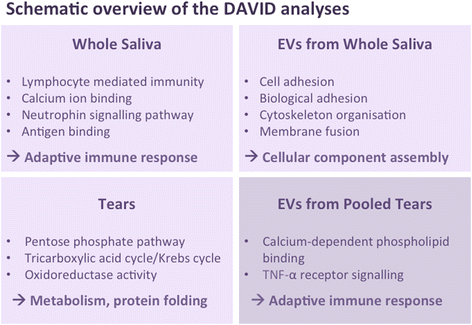
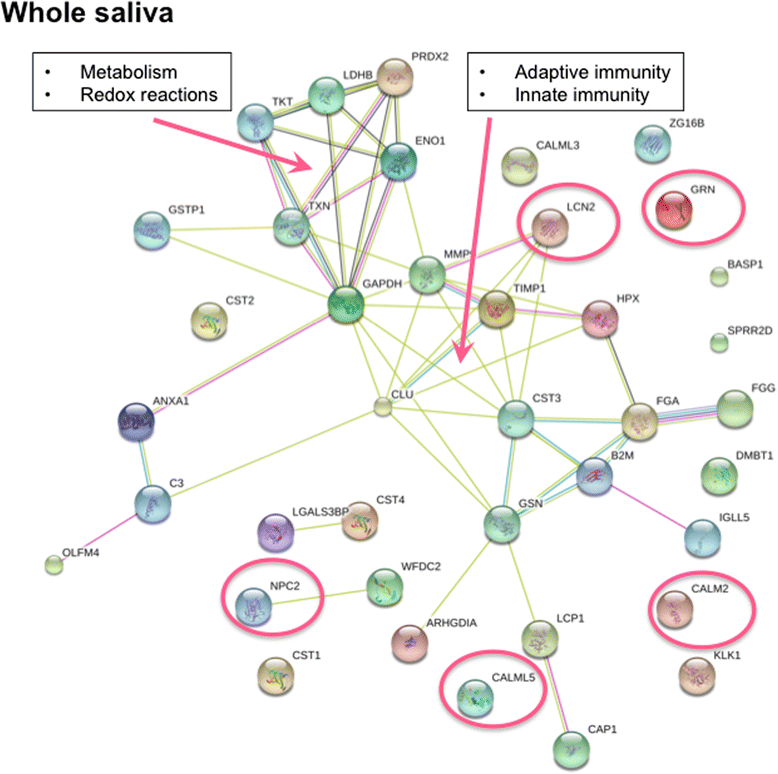
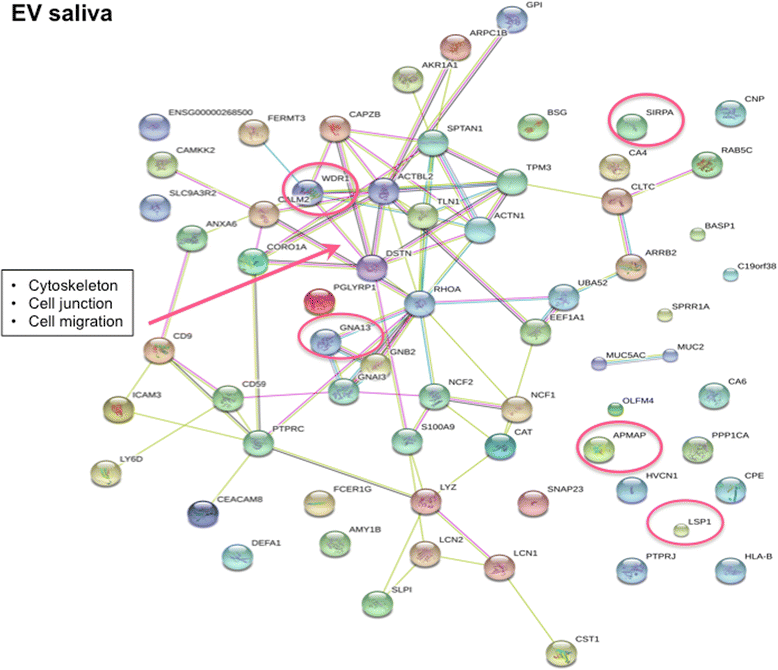
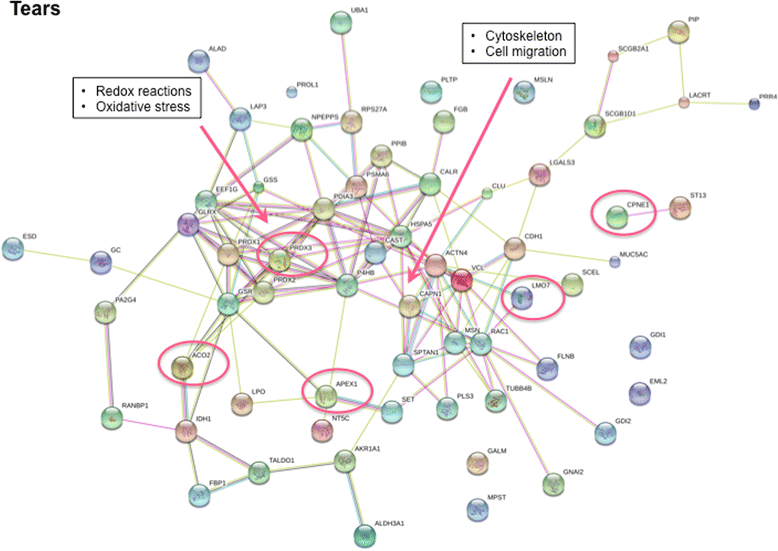
Results: Upregulation of proteins involved in innate immunity (LCN2), cell signalling (CALM) and wound repair (GRN and CALML5) were detected in saliva in pSS. Saliva EVs also displayed biomarkers critical for activation of the innate immune system (SIRPA and LSP1) and adipocyte differentiation (APMAP). Tear analysis indicated overexpression of proteins involved in TNF-α signalling (CPNE1) and B cell survival (PRDX3). Moreover, neutrophil gelatinase-associated lipocalin was upregulated in saliva and tears in pSS. Consistently, DAVID analysis demonstrated pathways of the adaptive immune response in saliva, of cellular component assembly for saliva EVs, and of metabolism and protein folding in tears in pSS patients.
Conclusions: LC-MS of saliva and tears from pSS patients, solely and in combination with size-exclusion chromatography allowed screening for possible novel biomarkers encompassing both salivary and lacrimal disease target organs. This approach could provide additional diagnostic accuracy in pSS, and could possibly also be applied for staging and monitoring the disease.
Keywords: Adaptive immunity; Autoimmunity; Biomarkers; Extracellular vesicles; Inflammation; Innate immunity; Proteomics; Saliva; Sjögren’s syndrome; Tears.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

