Localization of Protein Kinase NDR2 to Peroxisomes and Its Role in Ciliogenesis
- PMID: 28122914
- PMCID: PMC5354478
- DOI: 10.1074/jbc.M117.775916
Localization of Protein Kinase NDR2 to Peroxisomes and Its Role in Ciliogenesis
Abstract
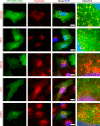
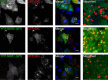
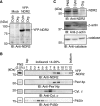
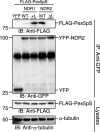
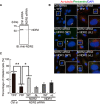
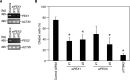
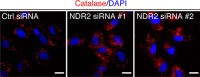
Nuclear Dbf2-related (NDR) kinases, comprising NDR1 and NDR2, are serine/threonine kinases that play crucial roles in the control of cell proliferation, apoptosis, and morphogenesis. We recently showed that NDR2, but not NDR1, is involved in primary cilium formation; however, the mechanism underlying their functional difference in ciliogenesis is unknown. To address this issue, we examined their subcellular localization. Despite their close sequence similarity, NDR2 exhibited punctate localization in the cytoplasm, whereas NDR1 was diffusely distributed within the cell. Notably, NDR2 puncta mostly co-localized with the peroxisome marker proteins, catalase and CFP-SKL (cyan fluorescent protein carrying the C-terminal typical peroxisome-targeting signal type-1 (PTS1) sequence, Ser-Lys-Leu). NDR2 contains the PTS1-like sequence, Gly-Lys-Leu, at the C-terminal end, whereas the C-terminal end of NDR1 is Ala-Lys. An NDR2 mutant lacking the C-terminal Leu, NDR2(ΔL), exhibited almost diffuse distribution in cells. Additionally, NDR2, but neither NDR1 nor NDR2(ΔL), bound to the PTS1 receptor Pex5p. Together, these findings indicate that NDR2 localizes to the peroxisome by using the C-terminal GKL sequence. Intriguingly, topology analysis of NDR2 suggests that NDR2 is exposed to the cytosolic surface of the peroxisome. The expression of wild-type NDR2, but not NDR2(ΔL), recovered the suppressive effect of NDR2 knockdown on ciliogenesis. Furthermore, knockdown of peroxisome biogenesis factor genes (PEX1 or PEX3) partially suppressed ciliogenesis. These results suggest that the peroxisomal localization of NDR2 is implicated in its function to promote primary cilium formation.
Keywords: NDR (nuclear dbf2-related) kinase; NDR2; cilia; ciliogenesis; peroisome targeting signal 1; peroxisome; protein kinase; protein targeting; protein translocation; serine/threonine protein kinase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Hergovich A., Stegert M. R., Schmitz D., and Hemmings B. A. (2006) NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7, 253–264 - PubMed
-
- Emoto K. (2011) The growing role of the Hippo-NDR kinase signaling in neuronal development and disease. J. Biochem. 150, 133–141 - PubMed
-
- Nagai T., and Mizuno K. (2014) Multifaceted roles of Furry proteins in invertebrates and vertebrates. J. Biochem. 155, 137–146 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

