Comparative Analysis of Mitochondrial N-Termini from Mouse, Human, and Yeast
- PMID: 28122942
- PMCID: PMC5383775
- DOI: 10.1074/mcp.M116.063818
Comparative Analysis of Mitochondrial N-Termini from Mouse, Human, and Yeast
Abstract
The majority of mitochondrial proteins are encoded in the nuclear genome, translated in the cytoplasm, and directed to the mitochondria by an N-terminal presequence that is cleaved upon import. Recently, N-proteome catalogs have been generated for mitochondria from yeast and from human U937 cells. Here, we applied the subtiligase method to determine N-termini for 327 proteins in mitochondria isolated from mouse liver and kidney. Comparative analysis between mitochondrial N-termini from mouse, human, and yeast proteins shows that whereas presequences are poorly conserved at the sequence level, other presequence properties are extremely conserved, including a length of ∼20-60 amino acids, a net charge between +3 to +6, and the presence of stabilizing amino acids at the N-terminus of mature proteins that follow the N-end rule from bacteria. As in yeast, ∼80% of mouse presequence cleavage sites match canonical motifs for three mitochondrial peptidases (MPP, Icp55, and Oct1), whereas the remainder do not match any known peptidase motifs. We show that mature mitochondrial proteins often exist with a spectrum of N-termini, consistent with a model of multiple cleavage events by MPP and Icp55. In addition to analysis of canonical targeting presequences, our N-terminal dataset allows the exploration of other cleavage events and provides support for polypeptide cleavage into two distinct enzymes (Hsd17b4), protein cleavages key for signaling (Oma1, Opa1, Htra2, Mavs, and Bcs2l13), and in several cases suggests novel protein isoforms (Scp2, Acadm, Adck3, Hsdl2, Dlst, and Ogdh). We present an integrated catalog of mammalian mitochondrial N-termini that can be used as a community resource to investigate individual proteins, to elucidate mechanisms of mammalian mitochondrial processing, and to allow researchers to engineer tags distally to the presequence cleavage.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
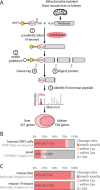
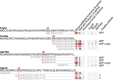
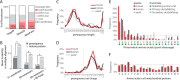
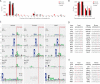

References
-
- Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., and Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 - PMC - PubMed
-
- Mossmann D., Meisinger C., and Vögtle F. N. (2012) Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 - PubMed
-
- Vögtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., and Meisinger C. (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

