Plant Actin-Depolymerizing Factors Possess Opposing Biochemical Properties Arising from Key Amino Acid Changes throughout Evolution
- PMID: 28123105
- PMCID: PMC5354190
- DOI: 10.1105/tpc.16.00690
Plant Actin-Depolymerizing Factors Possess Opposing Biochemical Properties Arising from Key Amino Acid Changes throughout Evolution
Abstract
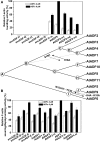
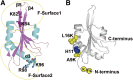
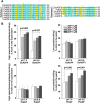
Functional divergence in paralogs is an important genetic source of evolutionary innovation. Actin-depolymerizing factors (ADFs) are among the most important actin binding proteins and are involved in generating and remodeling actin cytoskeletal architecture via their conserved F-actin severing or depolymerizing activity. In plants, ADFs coevolved with actin, but their biochemical properties are diverse. Unfortunately, the biochemical function of most plant ADFs and the potential mechanisms of their functional divergence remain unclear. Here, in vitro biochemical analyses demonstrated that all 11 ADF genes in Arabidopsis thaliana exhibit opposing biochemical properties. Subclass III ADFs evolved F-actin bundling (B-type) function from conserved F-actin depolymerizing (D-type) function, and subclass I ADFs have enhanced D-type function. By tracking historical mutation sites on ancestral proteins, several fundamental amino acid residues affecting the biochemical functions of these proteins were identified in Arabidopsis and various plants, suggesting that the biochemical divergence of ADFs has been conserved during the evolution of angiosperm plants. Importantly, N-terminal extensions on subclass III ADFs that arose from intron-sliding events are indispensable for the alteration of D-type to B-type function. We conclude that the evolution of these N-terminal extensions and several conserved mutations produced the diverse biochemical functions of plant ADFs from a putative ancestor.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures



Comment in
-
Loose-Knit Family: Tracing the Evolution of Actin-Depolymerizing Factors That Sever or Join the Actin Cytoskeleton.Plant Cell. 2017 Feb;29(2):200-201. doi: 10.1105/tpc.17.00088. Epub 2017 Jan 30. Plant Cell. 2017. PMID: 28138014 Free PMC article. No abstract available.
Similar articles
-
Research progress on the roles of actin-depolymerizing factor in plant stress responses.Front Plant Sci. 2023 Nov 16;14:1278311. doi: 10.3389/fpls.2023.1278311. eCollection 2023. Front Plant Sci. 2023. PMID: 38034575 Free PMC article. Review.
-
Higher-Ordered Actin Structures Remodeled by Arabidopsis ACTIN-DEPOLYMERIZING FACTOR5 Are Important for Pollen Germination and Pollen Tube Growth.Mol Plant. 2017 Aug 7;10(8):1065-1081. doi: 10.1016/j.molp.2017.06.001. Epub 2017 Jun 9. Mol Plant. 2017. PMID: 28606871
-
Nuclear Function of Subclass I Actin-Depolymerizing Factor Contributes to Susceptibility in Arabidopsis to an Adapted Powdery Mildew Fungus.Plant Physiol. 2016 Mar;170(3):1420-34. doi: 10.1104/pp.15.01265. Epub 2016 Jan 8. Plant Physiol. 2016. PMID: 26747284 Free PMC article.
-
Arabidopsis thaliana ACTIN DEPOLYMERIZING FACTORs play a role in leaf senescence regulation.Plant Cell Physiol. 2025 Jul 24;66(6):866-877. doi: 10.1093/pcp/pcaf027. Plant Cell Physiol. 2025. PMID: 40441715
-
Plant actin depolymerizing factor: actin microfilament disassembly and more.J Plant Res. 2017 Mar;130(2):227-238. doi: 10.1007/s10265-016-0899-8. Epub 2017 Jan 2. J Plant Res. 2017. PMID: 28044231 Free PMC article. Review.
Cited by
-
Genome-Wide Identification, Expression, and Interaction Analysis of the Auxin Response Factor and AUX/IAA Gene Families in Vaccinium bracteatum.Int J Mol Sci. 2024 Aug 1;25(15):8385. doi: 10.3390/ijms25158385. Int J Mol Sci. 2024. PMID: 39125955 Free PMC article.
-
Research progress on the roles of actin-depolymerizing factor in plant stress responses.Front Plant Sci. 2023 Nov 16;14:1278311. doi: 10.3389/fpls.2023.1278311. eCollection 2023. Front Plant Sci. 2023. PMID: 38034575 Free PMC article. Review.
-
Identification of R2R3-MYB family in blueberry and its potential involvement of anthocyanin biosynthesis in fruits.BMC Genomics. 2023 Aug 30;24(1):505. doi: 10.1186/s12864-023-09605-w. BMC Genomics. 2023. PMID: 37648968 Free PMC article.
-
Genome-wide characterization and expression analysis of the ADF gene family in response to salt and drought stress in alfalfa (Medicago sativa).Front Plant Sci. 2025 Jan 30;15:1520267. doi: 10.3389/fpls.2024.1520267. eCollection 2024. Front Plant Sci. 2025. PMID: 39949635 Free PMC article.
-
Heat-stable protein PGSL1 enhances pollen germination and tube growth at high temperature.Nat Commun. 2025 Apr 17;16(1):3642. doi: 10.1038/s41467-025-58869-1. Nat Commun. 2025. PMID: 40240780 Free PMC article.
References
-
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23. - PubMed
-
- Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J. (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94: 235–263. - PubMed
-
- Bowman G.D., Nodelman I.M., Hong Y., Chua N.H., Lindberg U., Schutt C.E. (2000). A comparative structural analysis of the ADF/cofilin family. Proteins 41: 374–384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

