The ruminal microbiome associated with methane emissions from ruminant livestock
- PMID: 28123698
- PMCID: PMC5244708
- DOI: 10.1186/s40104-017-0141-0
The ruminal microbiome associated with methane emissions from ruminant livestock
Abstract
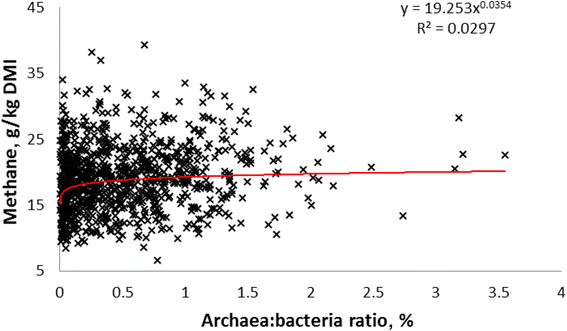
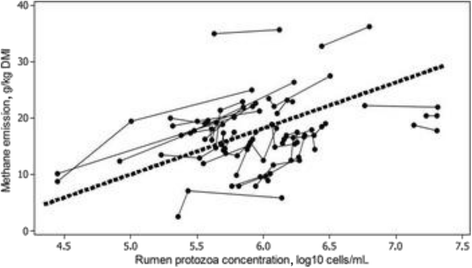
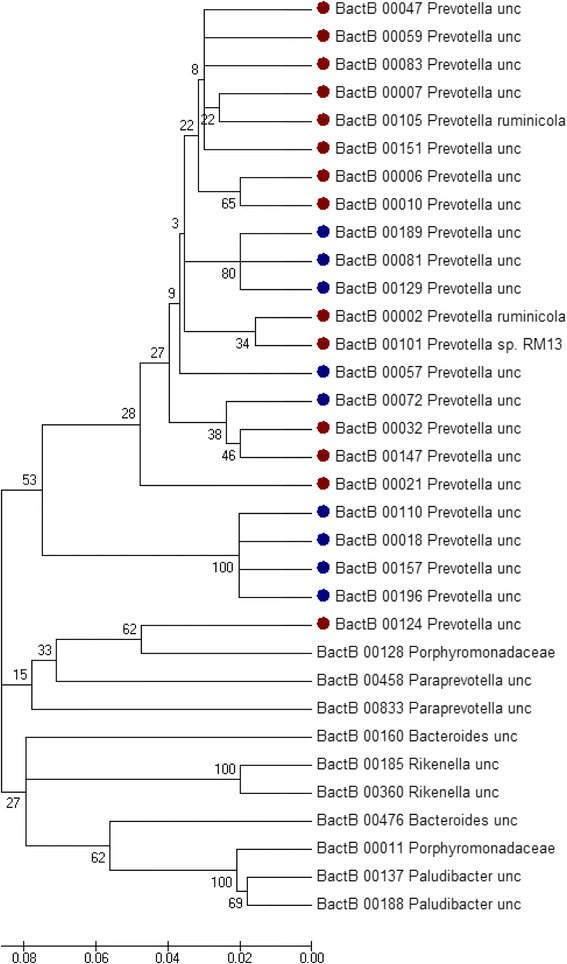
Methane emissions from ruminant livestock contribute significantly to the large environmental footprint of agriculture. The rumen is the principal source of methane, and certain features of the microbiome are associated with low/high methane phenotypes. Despite their primary role in methanogenesis, the abundance of archaea has only a weak correlation with methane emissions from individual animals. The composition of the archaeal community appears to have a stronger effect, with animals harbouring the Methanobrevibacter gottschalkii clade tending to be associated with greater methane emissions. Ciliate protozoa produce abundant H2, the main substrate for methanogenesis in the rumen, and their removal (defaunation) results in an average 11% lower methane emissions in vivo, but the results are not consistent. Different protozoal genera seem to result in greater methane emissions, though community types (A, AB, B and O) did not differ. Within the bacteria, three different 'ruminotypes' have been identified, two of which predispose animals to have lower methane emissions. The two low-methane ruminotypes are generally characterized by less abundant H2-producing bacteria. A lower abundance of Proteobacteria and differences in certain Bacteroidetes and anaerobic fungi seem to be associated with high methane emissions. Rumen anaerobic fungi produce abundant H2 and formate, and their abundance generally corresponds to the level of methane emissions. Thus, microbiome analysis is consistent with known pathways for H2 production and methanogenesis, but not yet in a predictive manner. The production and utilisation of formate by the ruminal microbiota is poorly understood and may be a source of variability between animals.
Keywords: Archaea; Methane; Microbiome; Rumen.
Figures
References
-
- IPCC . Climate change 2014: synthesis report. In: Pachauri RK, Meyer LA, editors. Contribution of working groups I, II and III to the fifth assessment report of the intergovernmental panel on climate change. Geneva: IPCC; 2014. p. 151.
-
- Hristov AN, Oh J, Lee C, Meinen R, Montes F, Ott F, et al. Mitigation of greenhouse gas emissions in livestock production. In: Gerber PJ, Henderson B, Makkar HPS, editors. A review of options for non-CO2 emissions. Rome: FAO; 2013. p. 226.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

