3'UTR AU-Rich Elements (AREs) and the RNA-Binding Protein Tristetraprolin (TTP) Are Not Required for the LPS-Mediated Destabilization of Phospholipase-Cβ-2 mRNA in Murine Macrophages
- PMID: 28124257
- PMCID: PMC5592789
- DOI: 10.1007/s10753-017-0511-y
3'UTR AU-Rich Elements (AREs) and the RNA-Binding Protein Tristetraprolin (TTP) Are Not Required for the LPS-Mediated Destabilization of Phospholipase-Cβ-2 mRNA in Murine Macrophages
Abstract
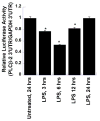
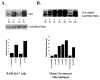
We have shown previously that bacterial lipopolysaccharide (LPS)-mediated suppression of phospholipase-Cβ-2 (PLCβ-2) expression is involved in M1 (inflammatory) to M2-like (wound healing) phenotypic switching of macrophages triggered by adenosine. This suppression is mediated post-transcriptionally by destabilization of PLCβ-2 mRNA (messenger ribonucleic acid). To investigate the mechanism of this LPS-mediated destabilization, we examined the roles of RNA-binding agents including microRNAs and RNA-binding proteins that are involved in regulating stability of mRNAs encoding growth factors, inflammatory mediators, and proto-oncogenes. Adenylate and uridylate (AU)-rich elements (AREs) in 3'UTRs are specific recognition sites for RNA-binding proteins including tristetraprolin (TTP), HuR, and AUF1 and for microRNAs that are involved in regulating mRNA stability. In this study, we investigated the role of TTP and AREs in regulating PLCβ-2 mRNA stability. The 3'UTR of the PLCβ-2 gene was inserted into the pLightswitch luciferase reporter plasmid and transfected into RAW264.7 cells. LPS suppressed luciferase expression from this reporter. Luciferase expression from mutant 3'UTR constructs lacking AREs was similarly downregulated, suggesting that these regions are not required for LPS-mediated suppression of PLCβ-2. TTP was rapidly upregulated in both primary murine macrophages and RAW264.7 cells in response to LPS. Suppression of PLCβ-2 by LPS was examined using macrophages from mice lacking TTP (TTP-/-). LPS suppressed PLCβ-2 expression to the same extent in wild type (WT) and TTP-/- macrophages. Also, the rate of decay of PLCβ-2 mRNA in LPS-treated macrophages following transcriptional blockade was similar in WT and TTP-/- macrophages, clearly indicating that TTP is not involved in LPS-mediated destabilization of PLCβ-2 mRNA in macrophages.
Keywords: 3’UTR; AU-rich elements; lipopolysaccharide (LPS); macrophages; phospholipase-Cβ-2; tristetraprolin.
Figures



References
-
- Goerdt S, Politz O, Schledzewski K, Birk R, Gratchev A, Guillot P, Hakiy N, Klemke CD, Dippel E, Kodelja V, Orfanos CE. Alternative versus classical activation of macrophages. Pathobiology. 1999;67:222–6. - PubMed
-
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. - PubMed
-
- Mantovani A, Bottazzi B, Sozzani S, Peri G, Allavena P, Dong QG, Vecchi A, Colotta F. Cytokine regulation of tumour-associated macrophages. Res Immunol. 1993;144:280–3. - PubMed
-
- Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

