Accessing gap-junction channel structure-function relationships through molecular modeling and simulations
- PMID: 28124624
- PMCID: PMC5267332
- DOI: 10.1186/s12860-016-0121-9
Accessing gap-junction channel structure-function relationships through molecular modeling and simulations
Abstract
Background: Gap junction channels (GJCs) are massive protein channels connecting the cytoplasm of adjacent cells. These channels allow intercellular transfer of molecules up to ~1 kDa, including water, ions and other metabolites. Unveiling structure-function relationships coded into the molecular architecture of these channels is necessary to gain insight on their vast biological function including electrical synapse, inflammation, development and tissular homeostasis. From early works, computational methods have been critical to analyze and interpret experimental observations. Upon the availability of crystallographic structures, molecular modeling and simulations have become a valuable tool to assess structure-function relationships in GJCs. Modeling different connexin isoforms, simulating the transport process, and exploring molecular variants, have provided new hypotheses and out-of-the-box approaches to the study of these important channels.
Methods: Here, we review foundational structural studies and recent developments on GJCs using molecular modeling and simulation techniques, highlighting the methods and the cross-talk with experimental evidence.
Results and discussion: By comparing results obtained by molecular modeling and simulations techniques with structural and functional information obtained from both recent literature and structural databases, we provide a critical assesment of structure-function relationships that can be obtained from the junction between theoretical and experimental evidence.
Keywords: Connexins; Gap-junction channels; Hemichannels; Homology modeling; Molecular simulation; Structure and function.
Figures
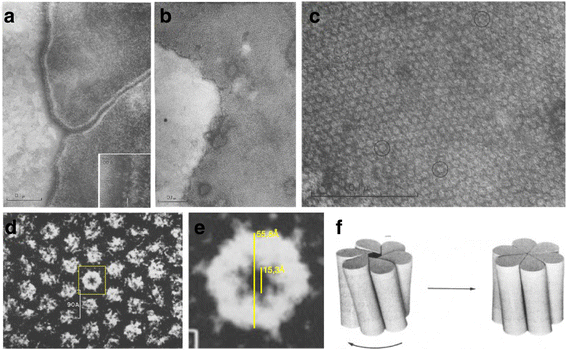




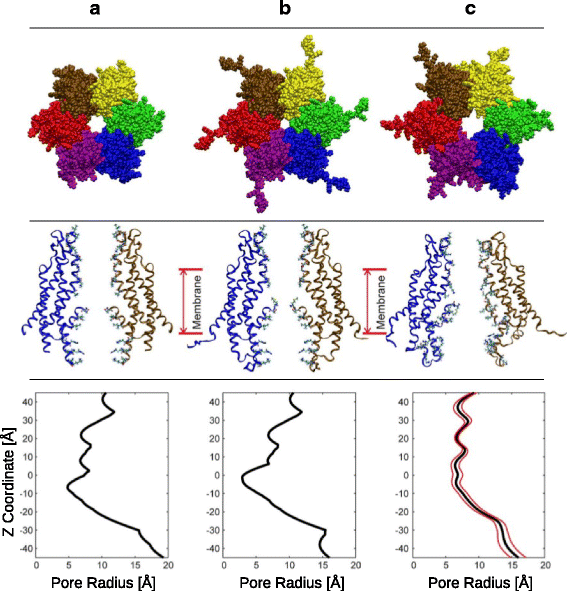
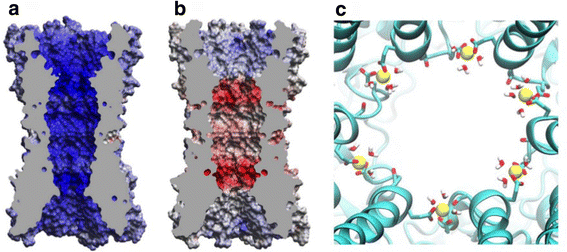

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

