Virus Escape and Manipulation of Cellular Nonsense-Mediated mRNA Decay
- PMID: 28124995
- PMCID: PMC5294993
- DOI: 10.3390/v9010024
Virus Escape and Manipulation of Cellular Nonsense-Mediated mRNA Decay
Abstract
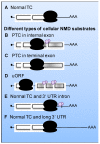
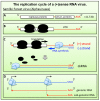
Nonsense-mediated mRNA decay (NMD), a cellular RNA turnover pathway targeting RNAs with features resulting in aberrant translation termination, has recently been found to restrict the replication of positive-stranded RNA ((+)RNA) viruses. As for every other antiviral immune system, there is also evidence of viruses interfering with and modulating NMD to their own advantage. This review will discuss our current understanding of why and how NMD targets viral RNAs, and elaborate counter-defense strategies viruses utilize to escape NMD.
Keywords: RNA quality control; RNA-protein interactions; gene expression; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

