Assessing the Genotypic Differences between Strains of Corynebacterium pseudotuberculosis biovar equi through Comparative Genomics
- PMID: 28125655
- PMCID: PMC5268413
- DOI: 10.1371/journal.pone.0170676
Assessing the Genotypic Differences between Strains of Corynebacterium pseudotuberculosis biovar equi through Comparative Genomics
Abstract
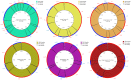
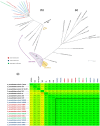
Seven genomes of Corynebacterium pseudotuberculosis biovar equi were sequenced on the Ion Torrent PGM platform, generating high-quality scaffolds over 2.35 Mbp. This bacterium is the causative agent of disease known as "pigeon fever" which commonly affects horses worldwide. The pangenome of biovar equi was calculated and two phylogenomic approaches were used to identify clustering patterns within Corynebacterium genus. Furthermore, other comparative analyses were performed including the prediction of genomic islands and prophages, and SNP-based phylogeny. In the phylogenomic tree, C. pseudotuberculosis was divided into two distinct clades, one formed by nitrate non-reducing species (biovar ovis) and another formed by nitrate-reducing species (biovar equi). In the latter group, the strains isolated from California were more related to each other, while the strains CIP 52.97 and 1/06-A formed the outermost clade of the biovar equi. A total of 1,355 core genes were identified, corresponding to 42.5% of the pangenome. This pangenome has one of the smallest core genomes described in the literature, suggesting a high genetic variability of biovar equi of C. pseudotuberculosis. The analysis of the similarity between the resistance islands identified a higher proximity between the strains that caused more severe infectious conditions (infection in the internal organs). Pathogenicity islands were largely conserved between strains. Several genes that modulate the pathogenicity of C. pseudotuberculosis were described including peptidases, recombination enzymes, micoside synthesis enzymes, bacteriocins with antimicrobial activity and several others. Finally, no genotypic differences were observed between the strains that caused the three different types of infection (external abscess formation, infection with abscess formation in the internal organs, and ulcerative lymphangitis). Instead, it was noted that there is a higher phenetic correlation between strains isolated at California compared to the other strains. Additionally, high variability of resistance islands suggests gene acquisition through several events of horizontal gene transfer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Aleman M, Spier SJ, Wilson WD, Doherr M. Retrospective study of Corynebacterium pseudotuberculosis infection in horses: 538 cases. J Am Vet Med Ass. 1996; 209: 804–809. - PubMed
-
- Spier SJ, Toth B, Edman J, Quave A, Habasha F, Garrick M, et al. Survival of Corynebacterium pseudotuberculosis biovar equi in soil. Vet Rec. 2012; 170(7): 180. - PubMed
-
- Spier SJ, Leutenegger CM, Carroll SP, Loye JE, Pusterla JB, Carpenter TE, et al. Use of a real-time polymerase chain reaction-based fluorogenic 5’ nuclease assay to evaluate insect vectors of Corynebacterium pseudotuberculosis infections in horses. Am J Vet Res. 2004; 65: 829–834. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

