Reversible Nuclear-Lipid-Droplet Morphology Induced by Oleic Acid: A Link to Cellular-Lipid Metabolism
- PMID: 28125673
- PMCID: PMC5268491
- DOI: 10.1371/journal.pone.0170608
Reversible Nuclear-Lipid-Droplet Morphology Induced by Oleic Acid: A Link to Cellular-Lipid Metabolism
Abstract
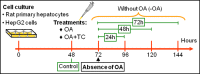
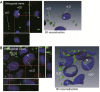
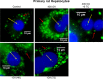
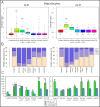
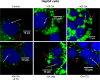
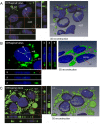
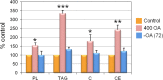
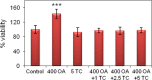
Neutral lipids-involved in many cellular processes-are stored as lipid droplets (LD), those mainly cytosolic (cLD) along with a small nuclear population (nLD). nLD could be involved in nuclear-lipid homeostasis serving as an endonuclear buffering system that would provide or incorporate lipids and proteins involved in signalling pathways as transcription factors and as enzymes of lipid metabolism and nuclear processes. Our aim was to determine if nLD constituted a dynamic domain. Oleic-acid (OA) added to rat hepatocytes or HepG2 cells in culture produced cellular-phenotypic LD modifications: increases in TAG, CE, C, and PL content and in cLD and nLD numbers and sizes. LD increments were reversed on exclusion of OA and were prevented by inhibition of acyl-CoA synthetase (with Triacsin C) and thus lipid biosynthesis. Under all conditions, nLD corresponded to a small population (2-10%) of total cellular LD. The anabolism triggered by OA, involving morphologic and size changes within the cLD and nLD populations, was reversed by a net balance of catabolism, upon eliminating OA. These catabolic processes included lipolysis and the mobilization of hydrolyzed FA from the LD to cytosolic-oxidation sites. These results would imply that nLD are actively involved in nuclear processes that include lipids. In conclusion, nLD are a dynamic nuclear domain since they are modified by OA through a reversible mechanism in combination with cLD; this process involves acyl-CoA-synthetase activity; ongoing TAG, CE, and PL biosynthesis. Thus, liver nLD and cLD are both dynamic cellular organelles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Farese R. V, & Walther T. C. (2016). Lipid droplets go nuclear. The Journal of Cell Biology, 212(1), 7–8. JOUR. Retrieved from http://jcb.rupress.org/content/212/1/7.abstract 10.1083/jcb.201512056 - DOI - PMC - PubMed
-
- Hillman N, Hillman R. Ultrastructural studies of tw32/tw32 mouse embryos. Development. 1975;33: 685–695. Available: http://dev.biologists.org/content/33/3/685.abstract - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

