The Number of Overlapping AID Hotspots in Germline IGHV Genes Is Inversely Correlated with Mutation Frequency in Chronic Lymphocytic Leukemia
- PMID: 28125682
- PMCID: PMC5268644
- DOI: 10.1371/journal.pone.0167602
The Number of Overlapping AID Hotspots in Germline IGHV Genes Is Inversely Correlated with Mutation Frequency in Chronic Lymphocytic Leukemia
Abstract
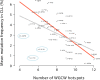
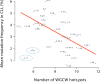
The targeting of mutations by Activation-Induced Deaminase (AID) is a key step in generating antibody diversity at the Immunoglobulin (Ig) loci but is also implicated in B-cell malignancies such as chronic lymphocytic leukemia (CLL). AID has previously been shown to preferentially deaminate WRC (W = A/T, R = A/G) hotspots. WGCW sites, which contain an overlapping WRC hotspot on both DNA strands, mutate at much higher frequency than single hotspots. Human Ig heavy chain (IGHV) genes differ in terms of WGCW numbers, ranging from 4 for IGHV3-48*03 to as many as 12 in IGHV1-69*01. An absence of V-region mutations in CLL patients ("IGHV unmutated", or U-CLL) is associated with a poorer prognosis compared to "IGHV mutated" (M-CLL) patients. The reasons for this difference are still unclear, but it has been noted that particular IGHV genes associate with U-CLL vs M-CLL. For example, patients with IGHV1-69 clones tend to be U-CLL with a poor prognosis, whereas patients with IGHV3-30 tend to be M-CLL and have a better prognosis. Another distinctive feature of CLL is that ~30% of (mostly poor prognosis) patients can be classified into "stereotyped" subsets, each defined by HCDR3 similarity, suggesting selection, possibly for a self-antigen. We analyzed >1000 IGHV genes from CLL patients and found a highly significant statistical relationship between the number of WGCW hotspots in the germline V-region and the observed mutation frequency in patients. However, paradoxically, this correlation was inverse, with V-regions with more WGCW hotspots being less likely to be mutated, i.e., more likely to be U-CLL. The number of WGCW hotspots in particular, are more strongly correlated with mutation frequency than either non-overlapping (WRC) hotspots or more general models of mutability derived from somatic hypermutation data. Furthermore, this correlation is not observed in sequences from the B cell repertoires of normal individuals and those with autoimmune diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. - PubMed
-
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–54. - PubMed
-
- Matthews C, Catherwood MA, Morris TC, Alexander HD. V(H)3-48 and V(H)3-53, as well as V(H)3-21, gene rearrangements define unique subgroups in CLL and are associated with biased lambda light chain restriction, homologous LCDR3 sequences and poor prognosis. Leuk Res. 2007;31(2):231–4. 10.1016/j.leukres.2006.03.028 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

