Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types
- PMID: 28126036
- PMCID: PMC5270224
- DOI: 10.1186/s13059-017-1156-8
Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types
Abstract
Background: A healthy immune system requires immune cells that adapt rapidly to environmental challenges. This phenotypic plasticity can be mediated by transcriptional and epigenetic variability.
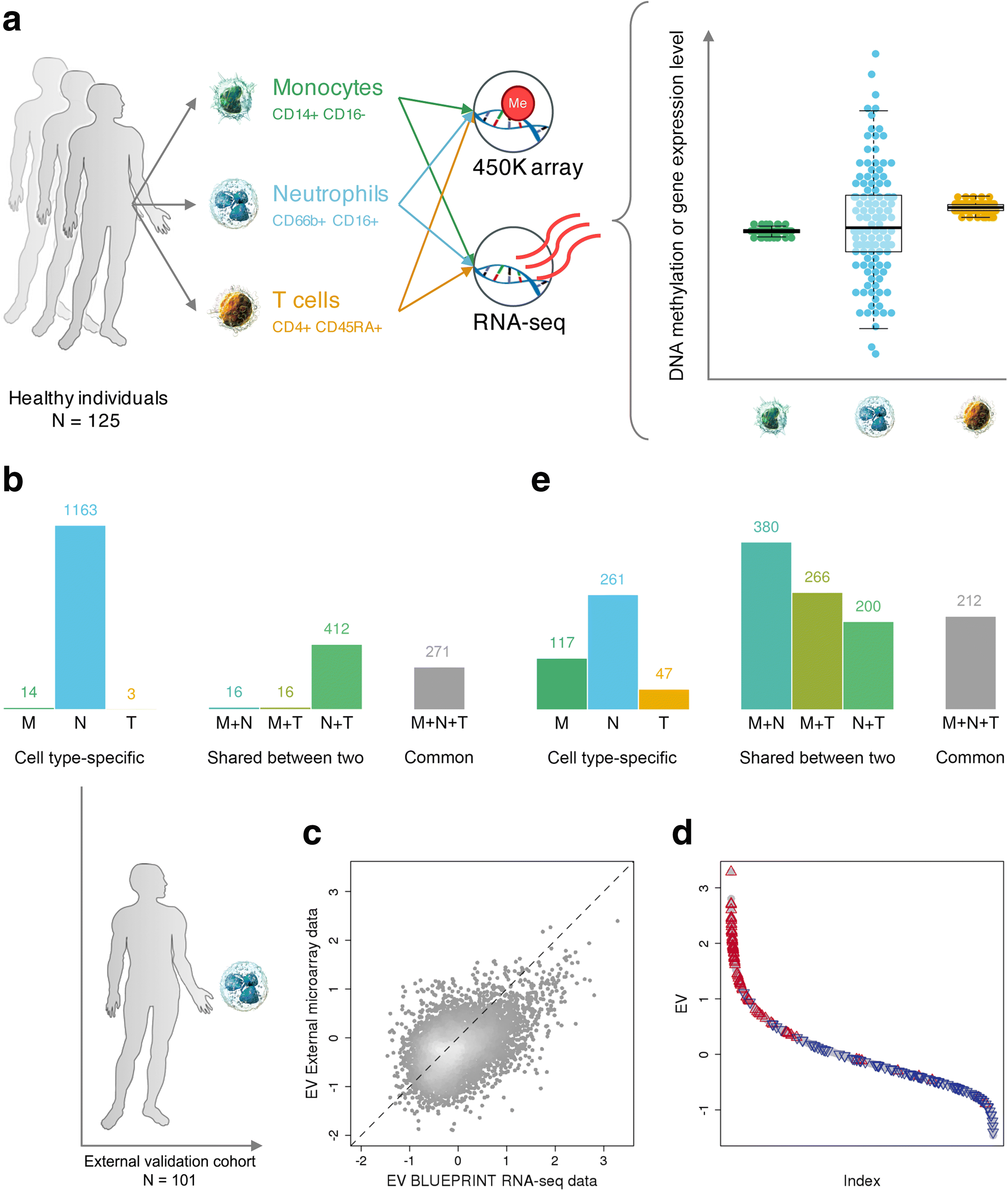
Results: We apply a novel analytical approach to measure and compare transcriptional and epigenetic variability genome-wide across CD14+CD16- monocytes, CD66b+CD16+ neutrophils, and CD4+CD45RA+ naïve T cells from the same 125 healthy individuals. We discover substantially increased variability in neutrophils compared to monocytes and T cells. In neutrophils, genes with hypervariable expression are found to be implicated in key immune pathways and are associated with cellular properties and environmental exposure. We also observe increased sex-specific gene expression differences in neutrophils. Neutrophil-specific DNA methylation hypervariable sites are enriched at dynamic chromatin regions and active enhancers.
Conclusions: Our data highlight the importance of transcriptional and epigenetic variability for the key role of neutrophils as the first responders to inflammatory stimuli. We provide a resource to enable further functional studies into the plasticity of immune cells, which can be accessed from: http://blueprint-dev.bioinfo.cnio.es/WP10/hypervariability .
Keywords: DNA methylation; Differential variability; Gene expression; Heterogeneity; Immune cells; Monocytes; Neutrophils; Phenotypic plasticity; T cells.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

