Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment
- PMID: 28126714
- PMCID: PMC5798883
- DOI: 10.1158/0008-5472.CAN-16-2490
Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment
Abstract
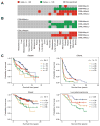
With the recent advent of immunotherapy, there is a critical need to understand immune cell interactions in the tumor microenvironment in both pan-cancer and tissue-specific contexts. Multidimensional datasets have enabled systematic approaches to dissect these interactions in large numbers of patients, furthering our understanding of the patient immune response to solid tumors. Using an integrated approach, we inferred the infiltration levels of distinct immune cell subsets in 23 tumor types from The Cancer Genome Atlas. From these quantities, we constructed a coinfiltration network, revealing interactions between cytolytic cells and myeloid cells in the tumor microenvironment. By integrating patient mutation data, we found that while mutation burden was associated with immune infiltration differences between distinct tumor types, additional factors likely explained differences between tumors originating from the same tissue. We concluded this analysis by examining the prognostic value of individual immune cell subsets as well as how coinfiltration of functionally discordant cell types associated with patient survival. In multiple tumor types, we found that the protective effect of CD8+ T cell infiltration was heavily modulated by coinfiltration of macrophages and other myeloid cell types, suggesting the involvement of myeloid-derived suppressor cells in tumor development. Our findings illustrate complex interactions between different immune cell types in the tumor microenvironment and indicate these interactions play meaningful roles in patient survival. These results demonstrate the importance of personalized immune response profiles when studying the factors underlying tumor immunogenicity and immunotherapy response. Cancer Res; 77(6); 1271-82. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
The authors declare they have no competing interests.
Figures



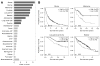
References
-
- Champiat S, Lambotte O, Barreau E, Belkhir R, Berdelou A, Carbonnel F, et al. Management of immune checkpoint blockade dysimmune toxicities: a collaborative position paper. Ann Oncol. 2016;27:559–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

