Identification of Phosphorylation Sites Altering Pollen Soluble Inorganic Pyrophosphatase Activity
- PMID: 28126844
- PMCID: PMC5338664
- DOI: 10.1104/pp.16.01450
Identification of Phosphorylation Sites Altering Pollen Soluble Inorganic Pyrophosphatase Activity
Abstract
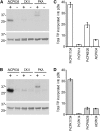
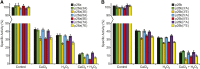
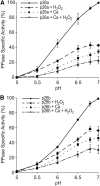
Protein phosphorylation regulates numerous cellular processes. Identifying the substrates and protein kinases involved is vital to understand how these important posttranslational modifications modulate biological function in eukaryotic cells. Pyrophosphatases catalyze the hydrolysis of inorganic phosphate (PPi) to inorganic phosphate Pi, driving biosynthetic reactions; they are essential for low cytosolic inorganic phosphate. It was suggested recently that posttranslational regulation of Family I soluble inorganic pyrophosphatases (sPPases) may affect their activity. We previously demonstrated that two pollen-expressed sPPases, Pr-p26.1a and Pr-p26.1b, from the flowering plant Papaver rhoeas were inhibited by phosphorylation. Despite the potential significance, there is a paucity of data on sPPase phosphorylation and regulation. Here, we used liquid chromatographic tandem mass spectrometry to map phosphorylation sites to the otherwise divergent amino-terminal extensions on these pollen sPPases. Despite the absence of reports in the literature on mapping phosphorylation sites on sPPases, a database survey of various proteomes identified a number of examples, suggesting that phosphorylation may be a more widely used mechanism to regulate these enzymes. Phosphomimetic mutants of Pr-p26.1a/b significantly and differentially reduced PPase activities by up to 2.5-fold at pH 6.8 and 52% in the presence of Ca2+ and hydrogen peroxide over unmodified proteins. This indicates that phosphoregulation of key sites can inhibit the catalytic responsiveness of these proteins in concert with key intracellular events. As sPPases are essential for many metabolic pathways in eukaryotic cells, our findings identify the phosphorylation of sPPases as a potential master regulatory mechanism that could be used to attenuate metabolism.
© 2017 The author(s). All Rights Reserved.
Figures



References
-
- Baykov AA, Avaeva SM (1981) A simple and sensitive apparatus for continuous monitoring of orthophosphate in the presence of acid-labile compounds. Anal Biochem 116: 1–4 - PubMed
-
- Buchanan B, Gruissem W, Jones R, editors (2002) Biochemistry and Molecular Biology of Plants. Wiley-Blackwell, New Jersey
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

