Effective number of breeders from sibship reconstruction: empirical evaluations using hatchery steelhead
- PMID: 28127391
- PMCID: PMC5253425
- DOI: 10.1111/eva.12433
Effective number of breeders from sibship reconstruction: empirical evaluations using hatchery steelhead
Abstract
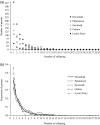
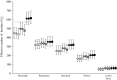
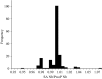
Effective population size (Ne ) is among the most important metrics in evolutionary biology. In natural populations, it is often difficult to collect adequate demographic data to calculate Ne directly. Consequently, genetic methods to estimate Ne have been developed. Two Ne estimators based on sibship reconstruction using multilocus genotype data have been developed in recent years: sibship assignment and parentage analysis without parents. In this study, we evaluated the accuracy of sibship reconstruction using a large empirical dataset from five hatchery steelhead populations with known pedigrees and using 95 single nucleotide polymorphism (SNP) markers. We challenged the software COLONY with 2,599,961 known relationships and demonstrated that reconstruction of full-sib and unrelated pairs was greater than 95% and 99% accurate, respectively. However, reconstruction of half-sib pairs was poor (<5% accurate). Despite poor half-sib reconstruction, both estimators provided accurate estimates of the effective number of breeders (Nb ) when sample sizes were near or greater than the true Nb and when assuming a monogamous mating system. We further demonstrated that both methods provide roughly equivalent estimates of Nb . Our results indicate that sibship reconstruction and current SNP panels provide promise for estimating Nb in steelhead populations in the region.
Keywords: COLONY; Oncorhynchus mykiss; PwoP; conservation genetics; effective population size; genetic monitoring; sibship assignment; sibship reconstruction.
Figures

References
-
- Abadía‐Cardoso, A. , Anderson, E. C. , Pearse, D. E. , & Garza, J. C. (2013). Large‐scale parentage analysis reveals reproductive patterns and heritability of spawn timing in a hatchery population of steelhead (Oncorhynchus mykiss). Molecular Ecology, 22, 4733–4746. - PubMed
-
- Anderson, E. C. 2010. Computational algorithms and user‐friendly software for parentage‐based tagging of Pacific salmonids [online]. Final report submitted to the Pacific Salmon Commission's Chinook Technical Committee (US Section). Retrieved from: https://swfsc.noaa.gov/uploadedFiles/Divisions/FED/Staff_Pages/Eric_Ande...
-
- Anderson, E. C. , & Ng, T. C. (2016). Bayesian pedigree inference with small numbers of single nucleotide polymorphisms via a factor‐graph representation. Theoretical Population Biology, 107, 39–51. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

