Transport capabilities of environmental Pseudomonads for sulfur compounds
- PMID: 28127814
- PMCID: PMC5368066
- DOI: 10.1002/pro.3124
Transport capabilities of environmental Pseudomonads for sulfur compounds
Abstract
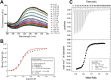
Sulfur is an essential element in plant rhizospheres and microbial activity plays a key role in increasing the biological availability of sulfur in soil environments. To better understand the mechanisms facilitating the exchange of sulfur-containing molecules in soil, we profiled the binding specificities of eight previously uncharacterized ABC transporter solute-binding proteins from plant-associated Pseudomonads. A high-throughput screening procedure indicated eighteen significant organosulfur binding ligands, with at least one high-quality screening hit for each protein target. Calorimetric and spectroscopic methods were used to validate the best ligand assignments and catalog the thermodynamic properties of the protein-ligand interactions. Two novel high-affinity ligand-binding activities were identified and quantified in this set of solute-binding proteins. Bacteria were cultured in minimal media with screening library components supplied as the sole sulfur sources, demonstrating that these organosulfur compounds can be metabolized and confirming the relevance of ligand assignments. These results expand the set of experimentally validated ligands amenable to transport by this ABC transporter family and demonstrate the complex range of protein-ligand interactions that can be accomplished by solute-binding proteins. Characterizing new nutrient import pathways provides insight into Pseudomonad metabolic capabilities which can be used to further interrogate bacterial survival and participation in soil and rhizosphere communities.
Keywords: ABC transporter; Pseudomonas fluorescens; Pseudomonas protegens; organosulfur compounds; rhizosphere.
© 2017 The Protein Society.
Figures


References
-
- Tetsch L, Jung K (2009) How are signals transduced across the cytoplasmic membrane? Transport proteins as transmitter of information. Amino Acids 37:467–477. - PubMed
-
- Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ (2009) Transport functions dominate the SAR11 metaproteome at low‐nutrient extremes in the Sargasso Sea. ISME J 3:93–105. - PubMed
-
- Tetsch L, Jung K (2009) The regulatory interplay between membrane‐integrated sensors and transport proteins in bacteria. Mol Microbiol 73:982–991. - PubMed
-
- Kertesz MA (2000) Riding the sulfur cycle–metabolism of sulfonates and sulfate esters in gram‐negative bacteria. FEMS Microbiol Rev 24:135–175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

