The IDL of E. coli SSB links ssDNA and protein binding by mediating protein-protein interactions
- PMID: 28127816
- PMCID: PMC5275737
- DOI: 10.1002/pro.3072
The IDL of E. coli SSB links ssDNA and protein binding by mediating protein-protein interactions
Abstract
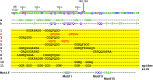
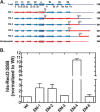
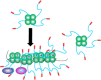
The E. coli single strand DNA binding protein (SSB) is essential to viability where it functions in two seemingly disparate roles: it binds to single stranded DNA (ssDNA) and to target proteins that comprise the SSB interactome. The link between these roles resides in a previously under-appreciated region of the protein known as the intrinsically disordered linker (IDL). We present a model wherein the IDL is responsible for mediating protein-protein interactions critical to each role. When interactions occur between SSB tetramers, cooperative binding to ssDNA results. When binding occurs between SSB and an interactome partner, storage or loading of that protein onto the DNA takes place. The properties of the IDL that facilitate these interactions include the presence of repeats, a putative polyproline type II helix and, PXXP motifs that may facilitate direct binding to the OB-fold in a manner similar to that observed for SH3 domain binding of PXXP ligands in eukaryotic systems.
Keywords: OB-fold; PXXP motif; RecG; RecO; SH3 domain; SSB.
© 2017 The Protein Society.
Figures





Similar articles
-
The intrinsically disordered linker of E. coli SSB is critical for the release from single-stranded DNA.Protein Sci. 2017 Apr;26(4):700-717. doi: 10.1002/pro.3115. Epub 2017 Mar 8. Protein Sci. 2017. PMID: 28078720 Free PMC article.
-
The mechanism of action of the SSB interactome reveals it is the first OB-fold family of genome guardians in prokaryotes.Protein Sci. 2021 Sep;30(9):1757-1775. doi: 10.1002/pro.4140. Epub 2021 Jun 14. Protein Sci. 2021. PMID: 34089559 Free PMC article. Review.
-
Are the intrinsically disordered linkers involved in SSB binding to accessory proteins?Nucleic Acids Res. 2019 Sep 19;47(16):8581-8594. doi: 10.1093/nar/gkz606. Nucleic Acids Res. 2019. PMID: 31329947 Free PMC article.
-
SSB and the RecG DNA helicase: an intimate association to rescue a stalled replication fork.Protein Sci. 2017 Apr;26(4):638-649. doi: 10.1002/pro.3114. Epub 2017 Mar 17. Protein Sci. 2017. PMID: 28078722 Free PMC article. Review.
-
The tale of SSB.Prog Biophys Mol Biol. 2017 Aug;127:111-118. doi: 10.1016/j.pbiomolbio.2016.11.001. Epub 2016 Nov 9. Prog Biophys Mol Biol. 2017. PMID: 27838363 Free PMC article. Review.
Cited by
-
The glycine-rich flexible region in SSB is crucial for PriA stimulation.RSC Adv. 2018 Oct 15;8(61):35280-35288. doi: 10.1039/c8ra07306f. eCollection 2018 Oct 10. RSC Adv. 2018. PMID: 35547063 Free PMC article.
-
Atomic force microscopy-based characterization of the interaction of PriA helicase with stalled DNA replication forks.J Biol Chem. 2020 May 1;295(18):6043-6052. doi: 10.1074/jbc.RA120.013013. Epub 2020 Mar 24. J Biol Chem. 2020. PMID: 32209655 Free PMC article.
-
The intrinsically disordered linker in the single-stranded DNA-binding protein influences DNA replication restart and recombination pathways in Escherichia coli K-12.J Bacteriol. 2024 Apr 18;206(4):e0033023. doi: 10.1128/jb.00330-23. Epub 2024 Mar 12. J Bacteriol. 2024. PMID: 38470036 Free PMC article.
-
Single-molecule insight into stalled replication fork rescue in Escherichia coli.Nucleic Acids Res. 2021 May 7;49(8):4220-4238. doi: 10.1093/nar/gkab142. Nucleic Acids Res. 2021. PMID: 33744948 Free PMC article. Review.
-
The Escherichia coli clamp loader rapidly remodels SSB on DNA to load clamps.Nucleic Acids Res. 2022 Dec 9;50(22):12872-12884. doi: 10.1093/nar/gkac1169. Nucleic Acids Res. 2022. PMID: 36511874 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

