Primary Human Placental Trophoblasts are Permissive for Zika Virus (ZIKV) Replication
- PMID: 28128342
- PMCID: PMC5269613
- DOI: 10.1038/srep41389
Primary Human Placental Trophoblasts are Permissive for Zika Virus (ZIKV) Replication
Abstract
Zika virus (ZIKV) is an emerging mosquito-borne (Aedes genus) arbovirus of the Flaviviridae family. Although ZIKV has been predominately associated with a mild or asymptomatic dengue-like disease, its appearance in the Americas has been accompanied by a multi-fold increase in reported incidence of fetal microcephaly and brain malformations. The source and mode of vertical transmission from mother to fetus is presumptively transplacental, although a causal link explaining the interval delay between maternal symptoms and observed fetal malformations following infection has been missing. In this study, we show that primary human placental trophoblasts from non-exposed donors (n = 20) can be infected by primary passage ZIKV-FLR isolate, and uniquely allowed for ZIKV viral RNA replication when compared to dengue virus (DENV). Consistent with their being permissive for ZIKV infection, primary trophoblasts expressed multiple putative ZIKV cell entry receptors, and cellular function and differentiation were preserved. These findings suggest that ZIKV-FLR strain can replicate in human placental trophoblasts without host cell destruction, thereby serving as a likely permissive reservoir and portal of fetal transmission with risk of latent microcephaly and malformations.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
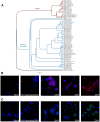
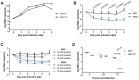

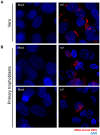
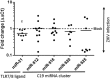
References
-
- Ioos S. et al. Current Zika virus epidemiology and recent epidemics. Med Mal Infect 44, 302–301 (2014). - PubMed
-
- Besnard M., Lastere S., Teissier A., Cao-Lormeau V. M. & Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill 19(13), pii = 20751 (2014). - PubMed
-
- Rapid risk assessment: Zika virus epidemic in the Americas: Potential association with microcephaly and Guillain-Barre syndrome. Stockholm: European Centre for Disease Prevention and Control, December 10, 2015 (http://ecdc.europa.eu/en/pulbications/zika-virus-americas-association-wi.... pdf).
-
- Ventura C. V., Maia M., Bravo-Fliho V., Gois A. L. & Belfort R. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387, 228 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

