Computational design of an epitope-specific Keap1 binding antibody using hotspot residues grafting and CDR loop swapping
- PMID: 28128368
- PMCID: PMC5269676
- DOI: 10.1038/srep41306
Computational design of an epitope-specific Keap1 binding antibody using hotspot residues grafting and CDR loop swapping
Abstract
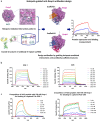
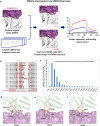
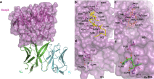
Therapeutic and diagnostic applications of monoclonal antibodies often require careful selection of binders that recognize specific epitopes on the target molecule to exert a desired modulation of biological function. Here we present a proof-of-concept application for the rational design of an epitope-specific antibody binding with the target protein Keap1, by grafting pre-defined structural interaction patterns from the native binding partner protein, Nrf2, onto geometrically matched positions of a set of antibody scaffolds. The designed antibodies bind to Keap1 and block the Keap1-Nrf2 interaction in an epitope-specific way. One resulting antibody is further optimised to achieve low-nanomolar binding affinity by in silico redesign of the CDRH3 sequences. An X-ray co-crystal structure of one resulting design reveals that the actual binding orientation and interface with Keap1 is very close to the design model, despite an unexpected CDRH3 tilt and VH/VL interface deviation, which indicates that the modelling precision may be improved by taking into account simultaneous CDR loops conformation and VH/VL orientation optimisation upon antibody sequence change. Our study confirms that, given a pre-existing crystal structure of the target protein-protein interaction, hotspots grafting with CDR loop swapping is an attractive route to the rational design of an antibody targeting a pre-selected epitope.
Conflict of interest statement
The authors declare competing financial interests: All authors are employees of UCB.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

