CRISPR/Cas9-Induced (CTG⋅CAG)n Repeat Instability in the Myotonic Dystrophy Type 1 Locus: Implications for Therapeutic Genome Editing
- PMID: 28129118
- PMCID: PMC5363205
- DOI: 10.1016/j.ymthe.2016.10.014
CRISPR/Cas9-Induced (CTG⋅CAG)n Repeat Instability in the Myotonic Dystrophy Type 1 Locus: Implications for Therapeutic Genome Editing
Abstract
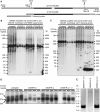
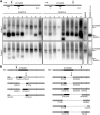
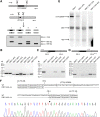
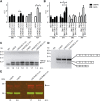
Myotonic dystrophy type 1 (DM1) is caused by (CTG⋅CAG)n-repeat expansion within the DMPK gene and thought to be mediated by a toxic RNA gain of function. Current attempts to develop therapy for this disease mainly aim at destroying or blocking abnormal properties of mutant DMPK (CUG)n RNA. Here, we explored a DNA-directed strategy and demonstrate that single clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-cleavage in either its 5' or 3' unique flank promotes uncontrollable deletion of large segments from the expanded trinucleotide repeat, rather than formation of short indels usually seen after double-strand break repair. Complete and precise excision of the repeat tract from normal and large expanded DMPK alleles in myoblasts from unaffected individuals, DM1 patients, and a DM1 mouse model could be achieved at high frequency by dual CRISPR/Cas9-cleavage at either side of the (CTG⋅CAG)n sequence. Importantly, removal of the repeat appeared to have no detrimental effects on the expression of genes in the DM1 locus. Moreover, myogenic capacity, nucleocytoplasmic distribution, and abnormal RNP-binding behavior of transcripts from the edited DMPK gene were normalized. Dual sgRNA-guided excision of the (CTG⋅CAG)n tract by CRISPR/Cas9 technology is applicable for developing isogenic cell lines for research and may provide new therapeutic opportunities for patients with DM1.
Keywords: (CTG⋅CAG)n repeat; CRISPR/Cas9; DM1 myoblasts; NHEJ; dsDNA break repair; myotonic dystrophy; somatic cell therapy; therapeutic genome editing; trinucleotide instability.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells.Nucleic Acids Res. 2018 Sep 19;46(16):8275-8298. doi: 10.1093/nar/gky548. Nucleic Acids Res. 2018. PMID: 29947794 Free PMC article.
-
Application of CRISPR-Cas9-Mediated Genome Editing for the Treatment of Myotonic Dystrophy Type 1.Mol Ther. 2020 Dec 2;28(12):2527-2539. doi: 10.1016/j.ymthe.2020.10.005. Epub 2020 Oct 14. Mol Ther. 2020. PMID: 33171139 Free PMC article. Review.
-
Genome Editing of Expanded CTG Repeats within the Human DMPK Gene Reduces Nuclear RNA Foci in the Muscle of DM1 Mice.Mol Ther. 2019 Aug 7;27(8):1372-1388. doi: 10.1016/j.ymthe.2019.05.021. Epub 2019 Jun 5. Mol Ther. 2019. PMID: 31253581 Free PMC article.
-
Muscle-specific gene editing improves molecular and phenotypic defects in a mouse model of myotonic dystrophy type 1.Clin Transl Med. 2025 Feb;15(2):e70227. doi: 10.1002/ctm2.70227. Clin Transl Med. 2025. PMID: 39956955 Free PMC article.
-
CRISPR/Cas Applications in Myotonic Dystrophy: Expanding Opportunities.Int J Mol Sci. 2019 Jul 27;20(15):3689. doi: 10.3390/ijms20153689. Int J Mol Sci. 2019. PMID: 31357652 Free PMC article. Review.
Cited by
-
CRISPR/Cas systems usher in a new era of disease treatment and diagnosis.Mol Biomed. 2022 Oct 14;3(1):31. doi: 10.1186/s43556-022-00095-y. Mol Biomed. 2022. PMID: 36239875 Free PMC article. Review.
-
Ameliorated cellular hallmarks of myotonic dystrophy in hybrid myotubes from patient and unaffected donor cells.Stem Cell Res Ther. 2024 Sep 15;15(1):302. doi: 10.1186/s13287-024-03913-y. Stem Cell Res Ther. 2024. PMID: 39278936 Free PMC article.
-
Establishment of quantitative and consistent in vitro skeletal muscle pathological models of myotonic dystrophy type 1 using patient-derived iPSCs.Sci Rep. 2023 Jan 11;13(1):94. doi: 10.1038/s41598-022-26614-z. Sci Rep. 2023. PMID: 36631509 Free PMC article.
-
Mapping the landscape of tandem repeat variability by targeted long read single molecule sequencing in familial X-linked intellectual disability.BMC Med Genomics. 2018 Dec 19;11(1):123. doi: 10.1186/s12920-018-0446-7. BMC Med Genomics. 2018. PMID: 30567555 Free PMC article.
-
Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells.Nucleic Acids Res. 2018 Sep 19;46(16):8275-8298. doi: 10.1093/nar/gky548. Nucleic Acids Res. 2018. PMID: 29947794 Free PMC article.
References
-
- Groenen P., Wieringa B. Expanding complexity in myotonic dystrophy. BioEssays. 1998;20:901–912. - PubMed
-
- Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
-
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O’Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

