Effects of control interventions on Clostridium difficile infection in England: an observational study
- PMID: 28130063
- PMCID: PMC5368411
- DOI: 10.1016/S1473-3099(16)30514-X
Effects of control interventions on Clostridium difficile infection in England: an observational study
Abstract
Background: The control of Clostridium difficile infections is an international clinical challenge. The incidence of C difficile in England declined by roughly 80% after 2006, following the implementation of national control policies; we tested two hypotheses to investigate their role in this decline. First, if C difficile infection declines in England were driven by reductions in use of particular antibiotics, then incidence of C difficile infections caused by resistant isolates should decline faster than that caused by susceptible isolates across multiple genotypes. Second, if C difficile infection declines were driven by improvements in hospital infection control, then transmitted (secondary) cases should decline regardless of susceptibility.
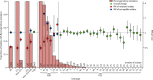
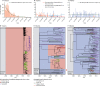
Methods: Regional (Oxfordshire and Leeds, UK) and national data for the incidence of C difficile infections and antimicrobial prescribing data (1998-2014) were combined with whole genome sequences from 4045 national and international C difficile isolates. Genotype (multilocus sequence type) and fluoroquinolone susceptibility were determined from whole genome sequences. The incidence of C difficile infections caused by fluoroquinolone-resistant and fluoroquinolone-susceptible isolates was estimated with negative-binomial regression, overall and per genotype. Selection and transmission were investigated with phylogenetic analyses.
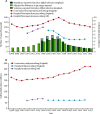
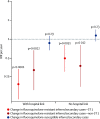
Findings: National fluoroquinolone and cephalosporin prescribing correlated highly with incidence of C difficile infections (cross-correlations >0·88), by contrast with total antibiotic prescribing (cross-correlations <0·59). Regionally, C difficile decline was driven by elimination of fluoroquinolone-resistant isolates (approximately 67% of Oxfordshire infections in September, 2006, falling to approximately 3% in February, 2013; annual incidence rate ratio 0·52, 95% CI 0·48-0·56 vs fluoroquinolone-susceptible isolates: 1·02, 0·97-1·08). C difficile infections caused by fluoroquinolone-resistant isolates declined in four distinct genotypes (p<0·01). The regions of phylogenies containing fluoroquinolone-resistant isolates were short-branched and geographically structured, consistent with selection and rapid transmission. The importance of fluoroquinolone restriction over infection control was shown by significant declines in inferred secondary (transmitted) cases caused by fluoroquinolone-resistant isolates with or without hospital contact (p<0·0001) versus no change in either group of cases caused by fluoroquinolone-susceptible isolates (p>0·2).
Interpretation: Restricting fluoroquinolone prescribing appears to explain the decline in incidence of C difficile infections, above other measures, in Oxfordshire and Leeds, England. Antimicrobial stewardship should be a central component of C difficile infection control programmes.
Funding: UK Clinical Research Collaboration (Medical Research Council, Wellcome Trust, National Institute for Health Research); NIHR Oxford Biomedical Research Centre; NIHR Health Protection Research Unit on Healthcare Associated Infection and Antimicrobial Resistance (Oxford University in partnership with Public Health England [PHE]), and on Modelling Methodology (Imperial College, London in partnership with PHE); and the Health Innovation Challenge Fund.
Copyright © 2017 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY license. Published by Elsevier Ltd.. All rights reserved.
Figures

Comment in
-
Fluoroquinolone restriction to control fluoroquinolone-resistant Clostridium difficile.Lancet Infect Dis. 2017 Apr;17(4):353-354. doi: 10.1016/S1473-3099(17)30052-X. Epub 2017 Jan 25. Lancet Infect Dis. 2017. PMID: 28130062 No abstract available.
-
Clostridium difficile in England: can we stop washing our hands? - Authors' reply.Lancet Infect Dis. 2017 May;17(5):478-479. doi: 10.1016/S1473-3099(17)30185-8. Lancet Infect Dis. 2017. PMID: 28447949 No abstract available.
-
Clostridium difficile in England: can we stop washing our hands?Lancet Infect Dis. 2017 May;17(5):478. doi: 10.1016/S1473-3099(17)30186-X. Lancet Infect Dis. 2017. PMID: 28447950 No abstract available.
References
-
- Davies KA, Longshaw CM, Davis GL. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14:1208–1219. - PubMed
-
- Owens RC, Jr, Donskey CJ, Gaynes RP, LooVG, Muto CA. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis. 2008;46:S19–S31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

