Melt With This Kiss: Paralyzing and Liquefying Venom of The Assassin Bug Pristhesancus plagipennis (Hemiptera: Reduviidae)
- PMID: 28130397
- PMCID: PMC5383778
- DOI: 10.1074/mcp.M116.063321
Melt With This Kiss: Paralyzing and Liquefying Venom of The Assassin Bug Pristhesancus plagipennis (Hemiptera: Reduviidae)
Abstract
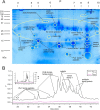
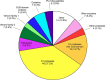
Assassin bugs (Hemiptera: Heteroptera: Reduviidae) are venomous insects, most of which prey on invertebrates. Assassin bug venom has features in common with venoms from other animals, such as paralyzing and lethal activity when injected, and a molecular composition that includes disulfide-rich peptide neurotoxins. Uniquely, this venom also has strong liquefying activity that has been hypothesized to facilitate feeding through the narrow channel of the proboscis-a structure inherited from sap- and phloem-feeding phytophagous hemipterans and adapted during the evolution of Heteroptera into a fang and feeding structure. However, further understanding of the function of assassin bug venom is impeded by the lack of proteomic studies detailing its molecular composition.By using a combined transcriptomic/proteomic approach, we show that the venom proteome of the harpactorine assassin bug Pristhesancus plagipennis includes a complex suite of >100 proteins comprising disulfide-rich peptides, CUB domain proteins, cystatins, putative cytolytic toxins, triabin-like protein, odorant-binding protein, S1 proteases, catabolic enzymes, putative nutrient-binding proteins, plus eight families of proteins without homology to characterized proteins. S1 proteases, CUB domain proteins, putative cytolytic toxins, and other novel proteins in the 10-16-kDa mass range, were the most abundant venom components. Thus, in addition to putative neurotoxins, assassin bug venom includes a high proportion of enzymatic and cytolytic venom components likely to be well suited to tissue liquefaction. Our results also provide insight into the trophic switch to blood-feeding by the kissing bugs (Reduviidae: Triatominae). Although some protein families such as triabins occur in the venoms of both predaceous and blood-feeding reduviids, the composition of venoms produced by these two groups is revealed to differ markedly. These results provide insights into the venom evolution in the insect suborder Heteroptera.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., and de la Vega R. C. (2009) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511 - PubMed
-
- Olivera B. M., Walker C., Cartier G. E., Hooper D., Santos A. D., Schoenfeld R., Shetty R., Watkins M., Bandyopadhyay P., and Hillyard D. R. (1999) Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Ann. N.Y. Acad. Sci. 870, 223–237 - PubMed
-
- Sollod B. L., Wilson D., Zhaxybayeva O., Gogarten J. P., Drinkwater R., and King G. F. (2005) Were arachnids the first to use combinatorial peptide libraries? Peptides 26, 131–139 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

