Extensive libraries of gene truncation variants generated by in vitro transposition
- PMID: 28130425
- PMCID: PMC5449547
- DOI: 10.1093/nar/gkx030
Extensive libraries of gene truncation variants generated by in vitro transposition
Abstract
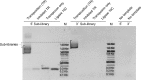
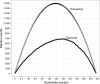
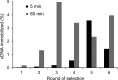
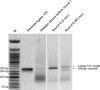
The detailed analysis of the impact of deletions on proteins or nucleic acids can reveal important functional regions and lead to variants with improved macromolecular properties. We present a method to generate large libraries of mutants with deletions of varying length that are randomly distributed throughout a given gene. This technique facilitates the identification of crucial sequence regions in nucleic acids or proteins. The approach utilizes in vitro transposition to generate 5΄ and 3΄ fragment sub-libraries of a given gene, which are then randomly recombined to yield a final library comprising both terminal and internal deletions. The method is easy to implement and can generate libraries in three to four days. We used this approach to produce a library of >9000 random deletion mutants of an artificial RNA ligase enzyme representing 32% of all possible deletions. The quality of the library was assessed by next-generation sequencing and detailed bioinformatics analysis. Finally, we subjected this library to in vitro selection and obtained fully functional variants with deletions of up to 18 amino acids of the parental enzyme that had been 95 amino acids in length.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Construction of block-shuffled libraries of DNA for evolutionary protein engineering: Y-ligation-based block shuffling.Protein Eng. 2002 Oct;15(10):843-53. doi: 10.1093/protein/15.10.843. Protein Eng. 2002. PMID: 12468719
-
Large-Scale Low-Cost NGS Library Preparation Using a Robust Tn5 Purification and Tagmentation Protocol.G3 (Bethesda). 2018 Jan 4;8(1):79-89. doi: 10.1534/g3.117.300257. G3 (Bethesda). 2018. PMID: 29118030 Free PMC article.
-
Easy preparation of a large-size random gene mutagenesis library in Escherichia coli.Anal Biochem. 2012 Sep 1;428(1):7-12. doi: 10.1016/j.ab.2012.05.022. Epub 2012 May 30. Anal Biochem. 2012. PMID: 22659340
-
Production of combinatorial libraries of fused genes by sequential transposition reactions.Nucleic Acids Res. 2002 Nov 1;30(21):e119. doi: 10.1093/nar/gnf118. Nucleic Acids Res. 2002. PMID: 12409478 Free PMC article.
-
Tn5 transposition: a molecular tool for studying protein structure-function.Biochem Soc Trans. 2006 Apr;34(Pt 2):320-3. doi: 10.1042/BST20060320. Biochem Soc Trans. 2006. PMID: 16545104 Review.
Cited by
-
Accessing unexplored regions of sequence space in directed enzyme evolution via insertion/deletion mutagenesis.Nat Commun. 2020 Jul 10;11(1):3469. doi: 10.1038/s41467-020-17061-3. Nat Commun. 2020. PMID: 32651386 Free PMC article.
-
Nature-inspired engineering of an artificial ligase enzyme by domain fusion.Nucleic Acids Res. 2022 Oct 28;50(19):11175-11185. doi: 10.1093/nar/gkac858. Nucleic Acids Res. 2022. PMID: 36243966 Free PMC article.
-
Enzyme miniaturization: Revolutionizing future biocatalysts.Biotechnol Adv. 2025 Sep;82:108598. doi: 10.1016/j.biotechadv.2025.108598. Epub 2025 May 10. Biotechnol Adv. 2025. PMID: 40354901 Free PMC article. Review.
-
Comprehensive deletion landscape of CRISPR-Cas9 identifies minimal RNA-guided DNA-binding modules.Nat Commun. 2021 Sep 27;12(1):5664. doi: 10.1038/s41467-021-25992-8. Nat Commun. 2021. PMID: 34580310 Free PMC article.
-
RanDeL-Seq: a High-Throughput Method to Map Viral cis- and trans-Acting Elements.mBio. 2021 Jan 19;12(1):e01724-20. doi: 10.1128/mBio.01724-20. mBio. 2021. PMID: 33468683 Free PMC article.
References
-
- Hecky J., Muller K.M.. Structural perturbation and compensation by directed evolution at physiological temperature leads to thermostabilization of β-lactamase. Biochemistry. 2005; 44:12640–12654. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

