Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma
- PMID: 28130639
- PMCID: PMC5600514
- DOI: 10.1007/s00401-017-1678-x
Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma
Abstract
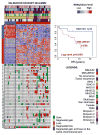
Meningioma is the most common primary brain tumor and carries a substantial risk of local recurrence. Methylation profiles of meningioma and their clinical implications are not well understood. We hypothesized that aggressive meningiomas have unique DNA methylation patterns that could be used to better stratify patient management. Samples (n = 140) were profiled using the Illumina HumanMethylation450BeadChip. Unsupervised modeling on a training set (n = 89) identified 2 molecular methylation subgroups of meningioma (MM) with significantly different recurrence-free survival (RFS) times between the groups: a prognostically unfavorable subgroup (MM-UNFAV) and a prognostically favorable subgroup (MM-FAV). This finding was validated in the remaining 51 samples and led to a baseline meningioma methylation classifier (bMMC) defined by 283 CpG loci (283-bMMC). To further optimize a recurrence predictor, probes subsumed within the baseline classifier were subject to additional modeling using a similar training/validation approach, leading to a 64-CpG loci meningioma methylation predictor (64-MMP). After adjustment for relevant clinical variables [WHO grade, mitotic index, Simpson grade, sex, location, and copy number aberrations (CNAs)] multivariable analyses for RFS showed that the baseline methylation classifier was not significant (p = 0.0793). The methylation predictor, however, was significantly associated with tumor recurrence (p < 0.0001). CNAs were extracted from the 450k intensity profiles. Tumor samples in the MM-UNFAV subgroup showed an overall higher proportion of CNAs compared to the MM-FAV subgroup tumors and the CNAs were complex in nature. CNAs in the MM-UNFAV subgroup included recurrent losses of 1p, 6q, 14q and 18q, and gain of 1q, all of which were previously identified as indicators of poor outcome. In conclusion, our analyses demonstrate robust DNA methylation signatures in meningioma that correlate with CNAs and stratify patients by recurrence risk.
Keywords: Copy number aberrations; DNA methylation; Epigenetics; Meningioma; Molecular subgroups; Recurrence risk.
Figures


References
-
- Abedalthagafi M, Bi WL, Aizer AA, Merrill PH, Brewster R, Agarwalla PK, Listewnik ML, Dias-Santagata D, Thorner AR, Van Hummelen P, Brastianos PK, Reardon DA, Wen PY, Al-Mefty O, Ramkissoon SH, Folkerth RD, Ligon KL, Ligon AH, Alexander BM, Dunn IF, Beroukhim R, Santagata S. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro-oncology. 2016;18(5):649–655. doi: 10.1093/neuonc/nov316. - DOI - PMC - PubMed
-
- Aghi MK, Carter BS, Cosgrove GR, Ojemann RG, Amin-Hanjani S, Martuza RL, Curry WT, Jr, Barker FG., 2nd Long-term recurrence rates of atypical meningiomas after gross total resection with or without postoperative adjuvant radiation. Neurosurgery. 2009;64(1):56–60. doi: 10.1227/01.NEU.0000330399.55586.63. discussion 60. - DOI - PubMed
-
- Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19(6):716–723.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

