Calpain-14 and its association with eosinophilic esophagitis
- PMID: 28131390
- PMCID: PMC5461191
- DOI: 10.1016/j.jaci.2016.09.027
Calpain-14 and its association with eosinophilic esophagitis
Abstract
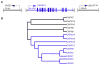
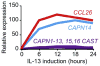
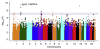
Calpains are a family of intracellular, calcium-dependent cysteine proteases involved in a variety of regulatory processes, including cytoskeletal dynamics, cell-cycle progression, signal transduction, gene expression, and apoptosis. These enzymes have been implicated in a number of disease processes, notably for this review involving eosinophilic tissue inflammation, such as eosinophilic esophagitis (EoE), a chronic inflammatory disorder triggered by allergic hypersensitivity to food and associated with genetic variants in calpain 14 (CAPN14). Herein we review the genetic, structural, and biochemical properties of CAPN14 and its gene product CAPN14, and its emerging role in patients with EoE. The CAPN14 gene is localized at chromosome 2p23.1-p21 and is most homologous to CAPN13 (36% sequence identity), which is located 365 kb downstream of CAPN14. Structurally, CAPN14 has classical calpain motifs, including a cysteine protease core. In comparison with other human calpains, CAPN14 has a unique expression pattern, with the highest levels in the upper gastrointestinal tract, particularly in the squamous epithelium of the esophagus. The CAPN14 gene is positioned in an epigenetic hotspot regulated by IL-13, a TH2 cytokine with increased levels in patients with EoE that has been shown to be a mediator of the disease. CAPN14 induces disruptive effects on the esophageal epithelium by impairing epithelial barrier function in association with loss of desmoglein-1 expression and has a regulatory role in repairing epithelial changes induced by IL-13. Thus CAPN14 is a unique protease with distinct tissue-specific expression and function in patients with EoE and is a potential therapeutic target for EoE and related eosinophilic and allergic diseases.
Keywords: Calpains; IL-13; calpain-14; calpainopathy; desmoglein-1; enzymes; eosinophilic esophagitis; eosinophils; epithelial barrier; food allergy; genetics; limb-girdle muscular dystrophy; myositis.
Copyright © 2017 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure of potential conflict of interest: L. C. Kottyan’s and M. E. Rothenberg’s institutions have received a grant from the National Institutes of Health. M. E. Rothenberg is a consultant for Immune Pharmaceuticals, NKT Therapeutics, Celgene, and Genetech and has an equity interest in the first two and receives royalties from Teva Pharmaceuticals for reslizumab. The rest of the authors declare that they have no relevant conflicts of interest.
Figures



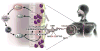
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

