Protein biomarker druggability profiling
- PMID: 28131723
- PMCID: PMC6286812
- DOI: 10.1016/j.jbi.2017.01.014
Protein biomarker druggability profiling
Abstract
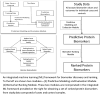
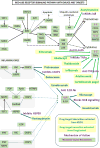
Developing automated and interactive methods for building a model by incorporating mechanistic and potentially causal annotations of ranked biomarkers of a disease or clinical condition followed by a mapping into a contextual framework in disease-linked biochemical pathways can be used for potential drug-target evaluation and for proposing new drug targets. We demonstrate the potential of this approach using ranked protein biomarkers obtained in neonatal sepsis by enrolling 127 infants (39 infants with late onset neonatal sepsis and 88 control infants) and by performing a focused proteomic profile of the sera and by applying the interactive druggability profiling algorithm (DPA) developed by us.
Keywords: Druggability profiling; Machine learning; Mechanistic annotation; Neonatal sepsis; Pathway analysis; Protein biomarkers.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

References
-
- Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. Journal of proteome research. 2005;4(4):1104–09. - PubMed
-
- Bogdanov M, Matson WR, Wang L, et al. Metabolomic profiling to develop blood biomarkers for Parkinson's disease. Brain. 2008;131(2):389–96. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

