DNA editing in DNA/RNA hybrids by adenosine deaminases that act on RNA
- PMID: 28132026
- PMCID: PMC5389660
- DOI: 10.1093/nar/gkx050
DNA editing in DNA/RNA hybrids by adenosine deaminases that act on RNA
Abstract
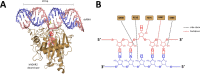
Adenosine deaminases that act on RNA (ADARs) carry out adenosine (A) to inosine (I) editing reactions with a known requirement for duplex RNA. Here, we show that ADARs also react with DNA/RNA hybrid duplexes. Hybrid substrates are deaminated efficiently by ADAR deaminase domains at dA-C mismatches and with E to Q mutations in the base flipping loop of the enzyme. For a long, perfectly matched hybrid, deamination is more efficient with full length ADAR2 than its isolated deaminase domain. Guide RNA strands for directed DNA editing by ADAR were used to target six different 2΄-deoxyadenosines in the M13 bacteriophage ssDNA genome. DNA editing efficiencies varied depending on the sequence context of the editing site consistent with known sequence preferences for ADARs. These observations suggest the reaction within DNA/RNA hybrids may be a natural function of human ADARs. In addition, this work sets the stage for development of a new class of genome editing tools based on directed deamination of 2΄-deoxyadenosines in DNA/RNA hybrids.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs.Bioessays. 2017 Apr;39(4):10.1002/bies.201600187. doi: 10.1002/bies.201600187. Epub 2017 Feb 20. Bioessays. 2017. PMID: 28217931 Free PMC article. Review.
-
Adenosine Deaminases That Act on RNA (ADARs).Enzymes. 2017;41:215-268. doi: 10.1016/bs.enz.2017.03.006. Epub 2017 Apr 14. Enzymes. 2017. PMID: 28601223 Review.
-
Effect of mismatch on binding of ADAR2/GluR-2 pre-mRNA complex.J Mol Model. 2015 Sep;21(9):222. doi: 10.1007/s00894-015-2760-8. Epub 2015 Aug 8. J Mol Model. 2015. PMID: 26252972
-
Base-pairing probability in the microRNA stem region affects the binding and editing specificity of human A-to-I editing enzymes ADAR1-p110 and ADAR2.RNA Biol. 2018;15(7):976-989. doi: 10.1080/15476286.2018.1486658. Epub 2018 Jul 24. RNA Biol. 2018. PMID: 29950133 Free PMC article.
-
Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.Nat Struct Mol Biol. 2016 May;23(5):426-33. doi: 10.1038/nsmb.3203. Epub 2016 Apr 11. Nat Struct Mol Biol. 2016. PMID: 27065196 Free PMC article.
Cited by
-
Engineered deaminases as a key component of DNA and RNA editing tools.Mol Ther Nucleic Acids. 2023 Oct 20;34:102062. doi: 10.1016/j.omtn.2023.102062. eCollection 2023 Dec 12. Mol Ther Nucleic Acids. 2023. PMID: 38028200 Free PMC article. Review.
-
Predicting clinical outcomes using cancer progression associated signatures.Oncotarget. 2021 Apr 13;12(8):845-858. doi: 10.18632/oncotarget.27934. eCollection 2021 Apr 13. Oncotarget. 2021. PMID: 33889305 Free PMC article.
-
Evolutionary honing in and mutational replacement: how long-term directed mutational responses to specific environmental pressures are possible.Theory Biosci. 2023 Jun;142(2):87-105. doi: 10.1007/s12064-023-00387-z. Epub 2023 Mar 11. Theory Biosci. 2023. PMID: 36899155 Free PMC article. Review.
-
In What Ways Do Synthetic Nucleotides and Natural Base Lesions Alter the Structural Stability of G-Quadruplex Nucleic Acids?J Nucleic Acids. 2017;2017:1641845. doi: 10.1155/2017/1641845. Epub 2017 Oct 18. J Nucleic Acids. 2017. PMID: 29181193 Free PMC article. Review.
-
Inosine in Biology and Disease.Genes (Basel). 2021 Apr 19;12(4):600. doi: 10.3390/genes12040600. Genes (Basel). 2021. PMID: 33921764 Free PMC article. Review.
References
-
- Bass B.L., Weintraub H.. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988; 55:1089–1098. - PubMed
-
- Goodman R.A., Macbeth M.R., Beal P.A.. Samuel CE. Adenosine Deaminases Acting on RNA (ADARs) and A-to-I Editing. 2012; Berlin, Heidelberg: Springer Berlin Heidelberg; 1–33.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

